Deposition Date
2000-04-05
Release Date
2001-01-16
Last Version Date
2024-05-15
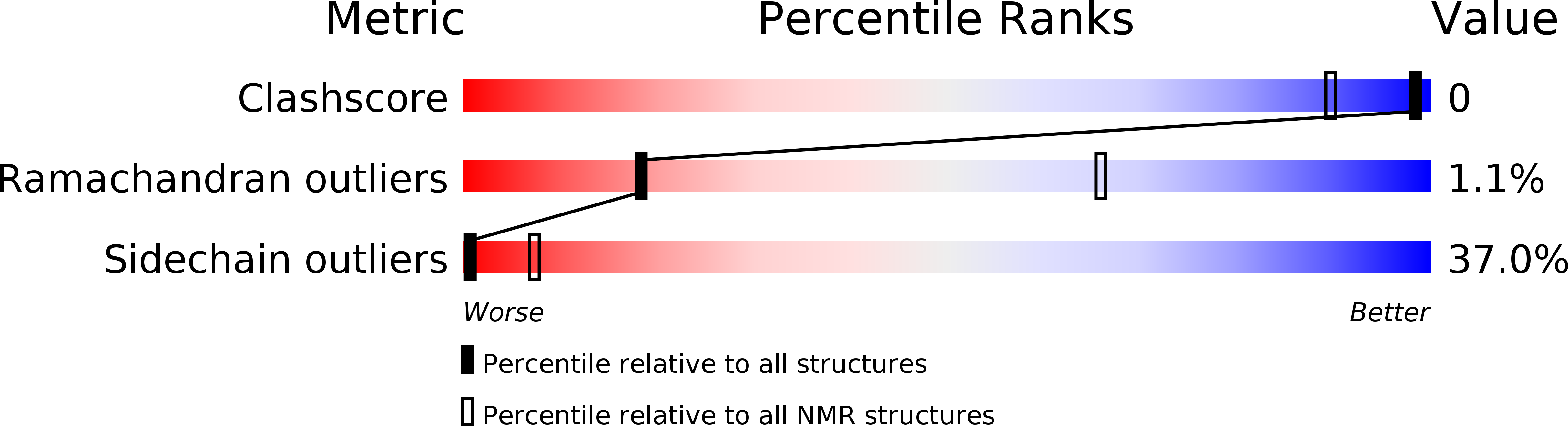
Entry Detail
PDB ID:
1E0Q
Keywords:
Title:
Mutant Peptide from the first N-terminal 17 amino-acid of Ubiquitin
Biological Source:
Source Organism(s):
BOS TAURUS (Taxon ID: 9913)
Method Details:
Experimental Method:
Conformers Calculated:
60
Conformers Submitted:
27
Selection Criteria:
LEAST RESTRAINT VIOLATION