Deposition Date
2000-01-11
Release Date
2000-07-26
Last Version Date
2024-04-10
Entry Detail
PDB ID:
1DT7
Keywords:
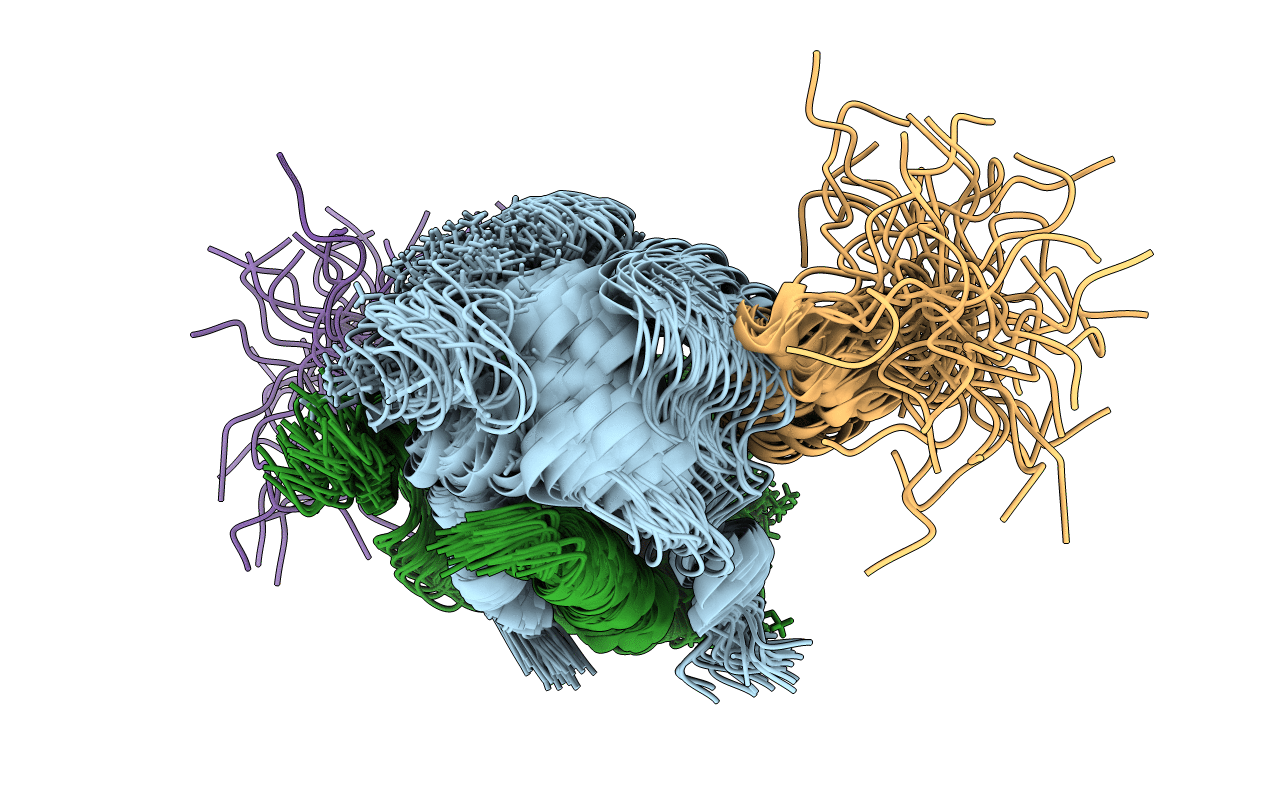
Title:
SOLUTION STRUCTURE OF THE C-TERMINAL NEGATIVE REGULATORY DOMAIN OF P53 IN A COMPLEX WITH CA2+-BOUND S100B(BB)
Biological Source:
Source Organism(s):
Rattus norvegicus (Taxon ID: 10116)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
40
Conformers Submitted:
40
Selection Criteria:
structure with the lowest energy and the least restraint violations


