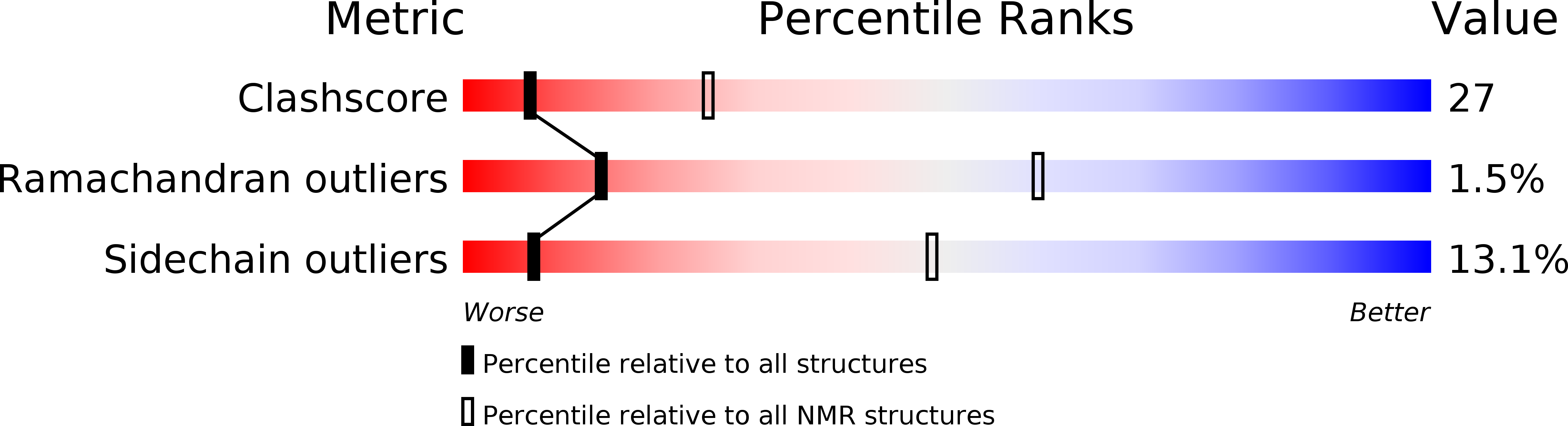
Deposition Date
1999-12-06
Release Date
2000-02-14
Last Version Date
2024-04-10
Entry Detail
PDB ID:
1DK2
Keywords:
Title:
REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA
Biological Source:
Source Organism(s):
Rattus norvegicus (Taxon ID: 10116)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
25
Selection Criteria:
THE SELECTION UTILIZED THE LOWEST ENERGY STRUCTURES WITH NO NOE VIOLATIONS
EXCEEDING 0.3 ANGSTROMS AND NO DIHEDRAL VIOLATIONS EXCEEDING 0.3 DEGREES.