Deposition Date
1999-11-04
Release Date
2000-08-23
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1DC3
Keywords:
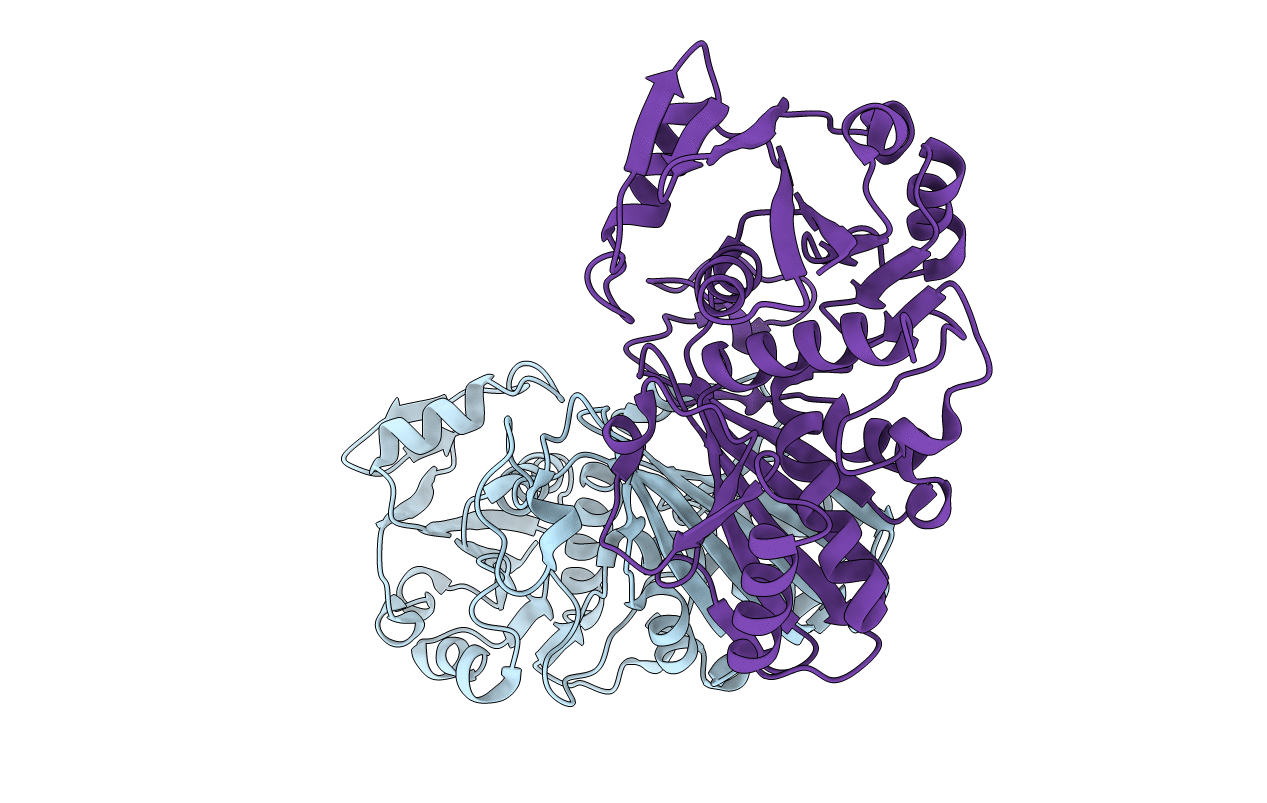
Title:
STRUCTURAL ANALYSIS OF GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE FROM ESCHERICHIA COLI: DIRECT EVIDENCE FOR SUBSTRATE BINDING AND COFACTOR-INDUCED CONFORMATIONAL CHANGES
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Method Details:
Experimental Method:
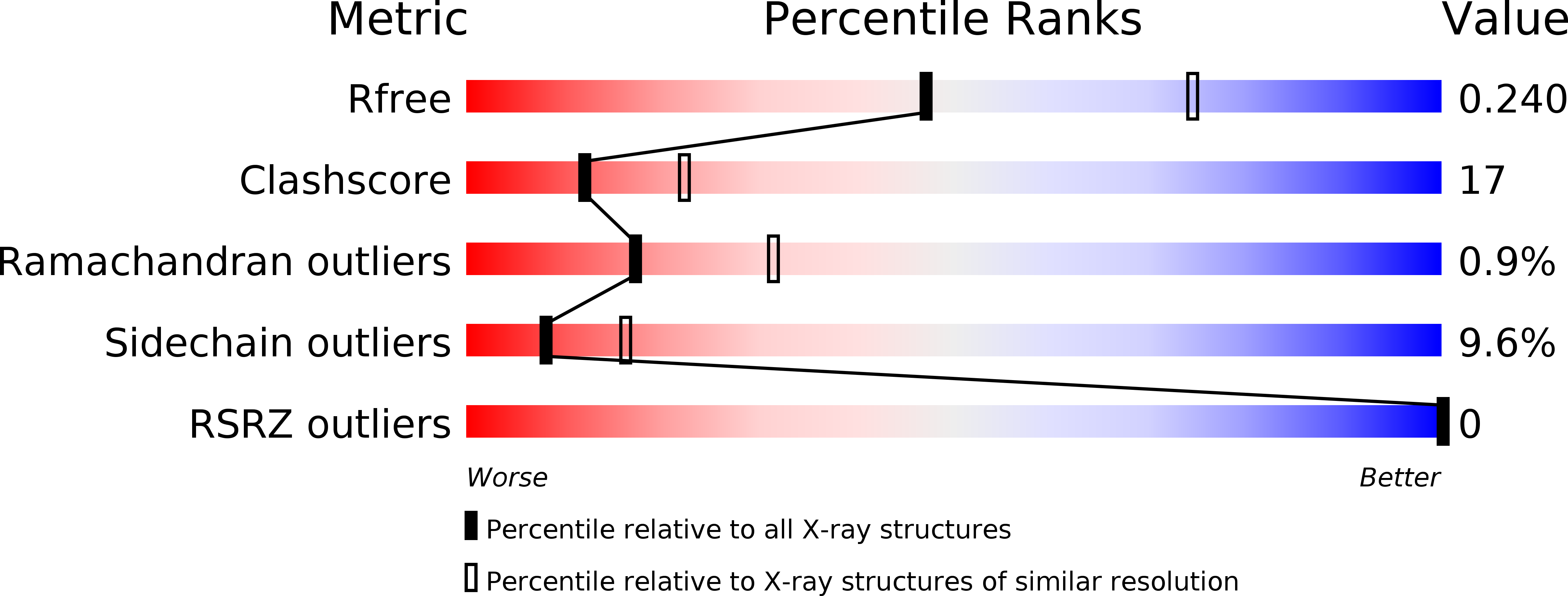
Resolution:
2.50 Å
R-Value Free:
0.24
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
I 41