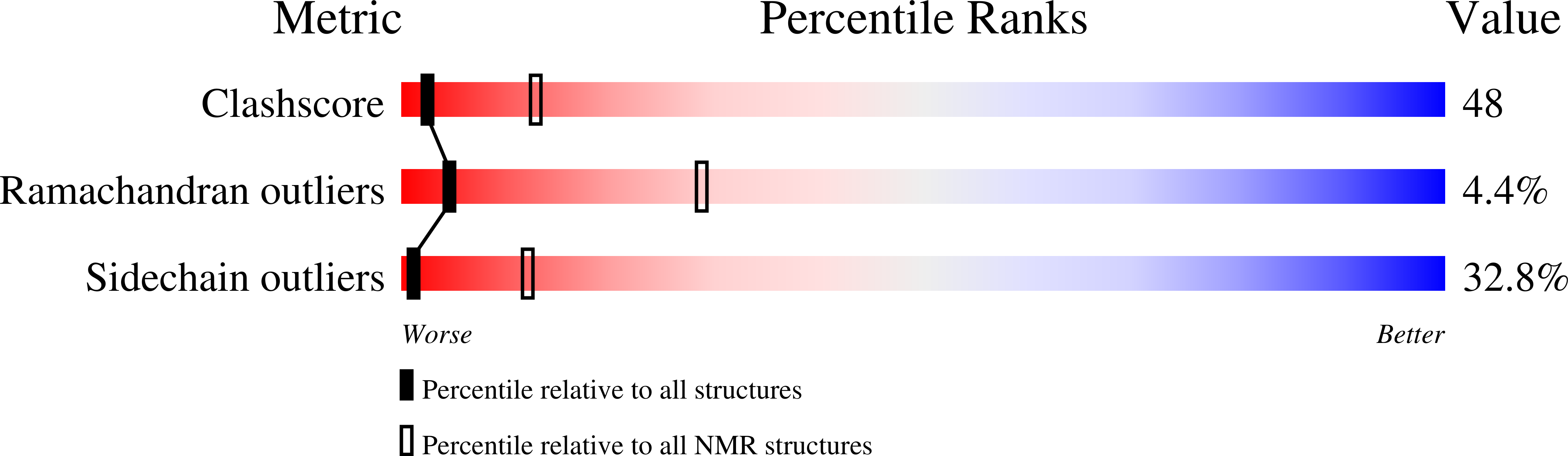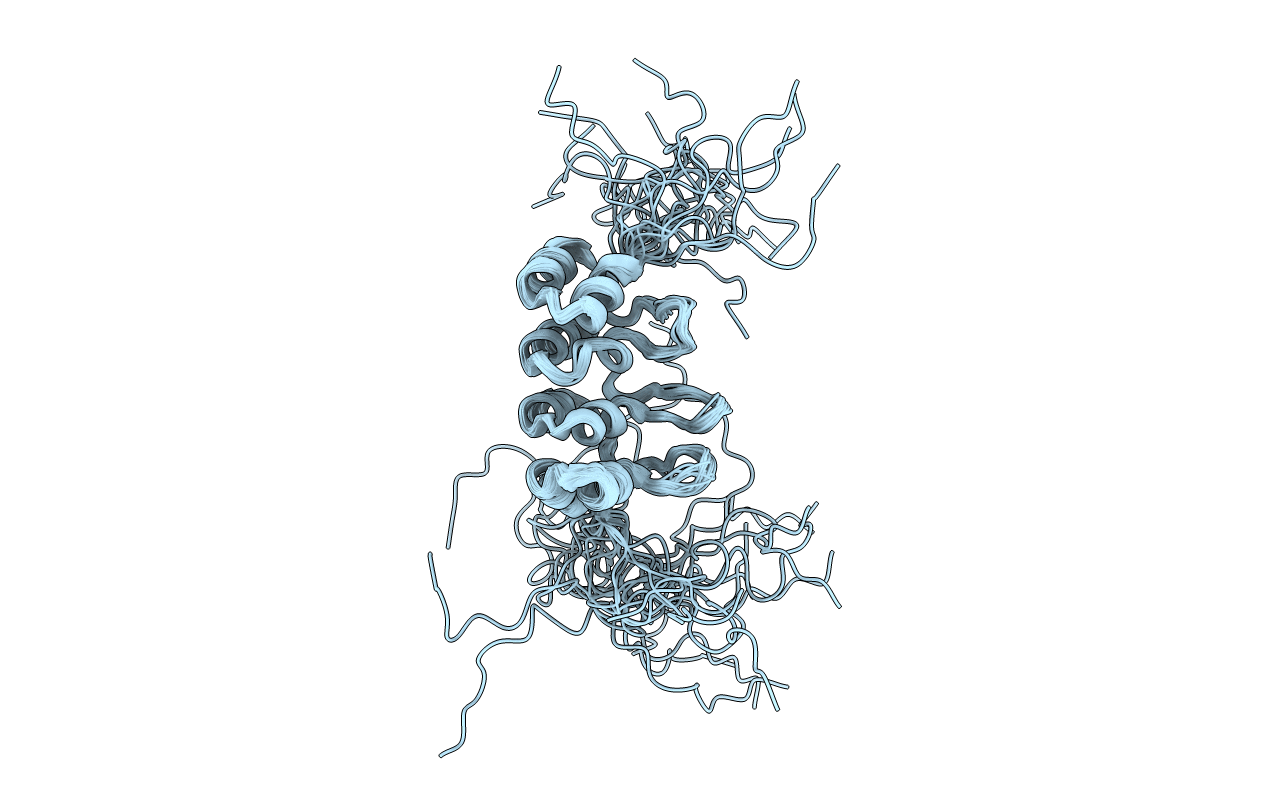
Deposition Date
1999-11-04
Release Date
1999-12-23
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1DC2
Keywords:
Title:
SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 20 STRUCTURES
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
60
Conformers Submitted:
20
Selection Criteria:
THE CLOSEST TO MEAN STRUCTURE WHICH SHOWS GOOD AGREEMENT WITH THE EXPERIMENTAL
RESTRAINTS