Deposition Date
1997-12-01
Release Date
1999-01-13
Last Version Date
2024-05-22
Entry Detail
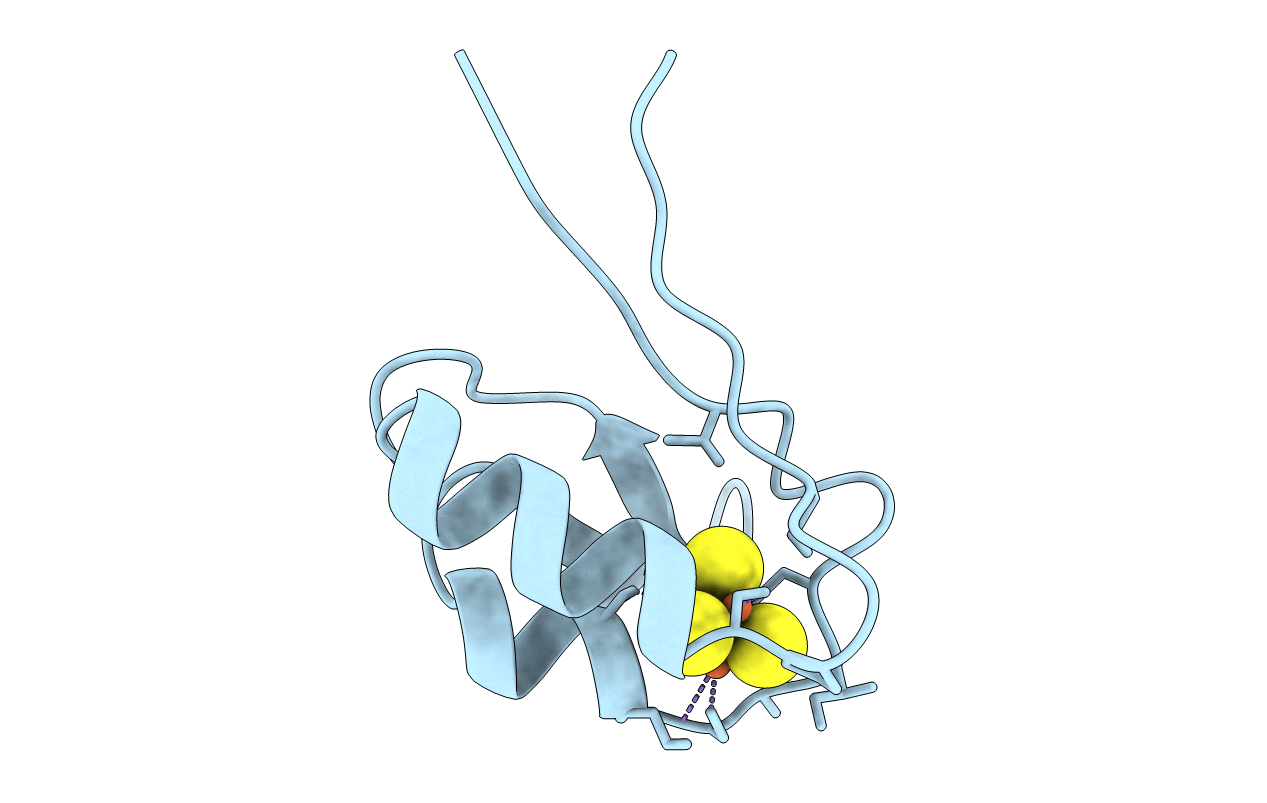
PDB ID:
1DAX
Keywords:
Title:
OXIDISED DESULFOVIBRIO AFRICANUS FERREDOXIN I, NMR, MINIMIZED AVERAGE STRUCTURE
Biological Source:
Source Organism(s):
Desulfovibrio africanus (Taxon ID: 873)
Method Details:


