Deposition Date
1999-10-19
Release Date
1999-11-19
Last Version Date
2023-08-09
Entry Detail
PDB ID:
1D7U
Keywords:
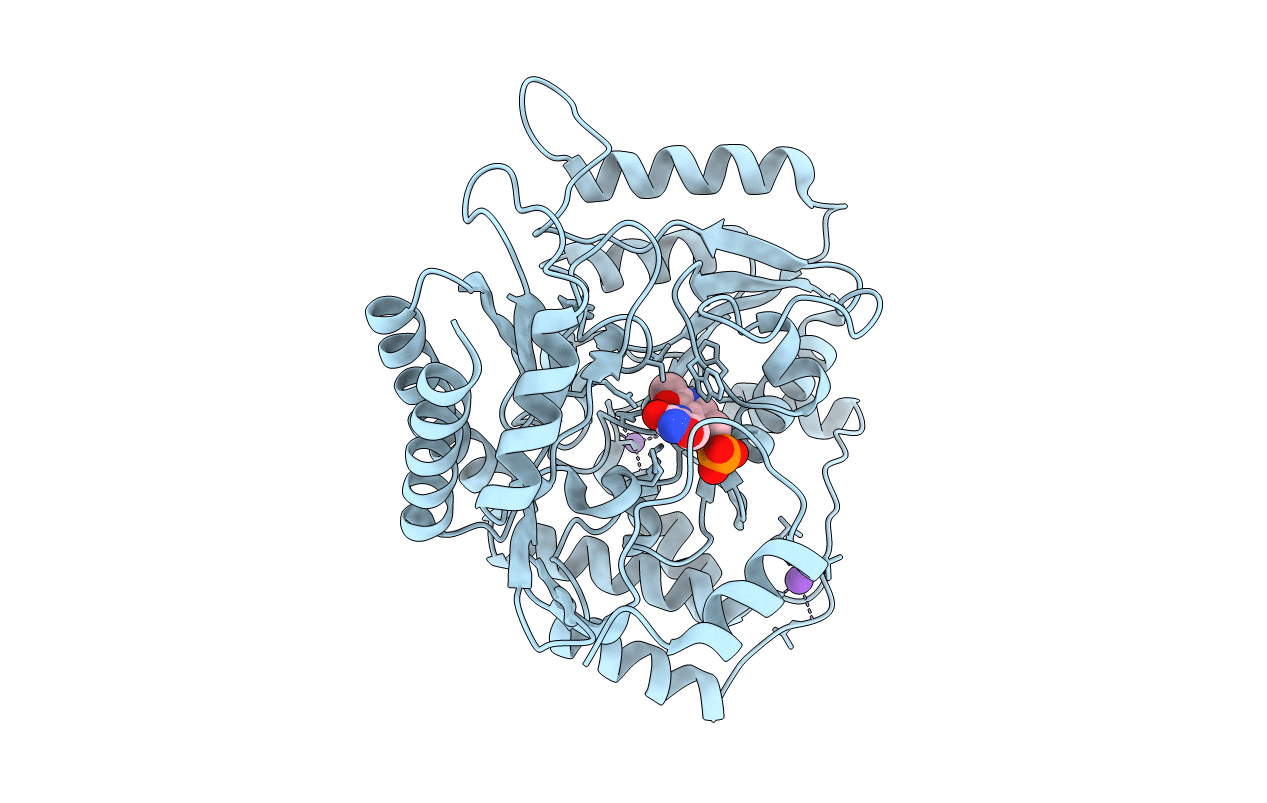
Title:
Crystal structure of the complex of 2,2-dialkylglycine decarboxylase with LCS
Biological Source:
Source Organism(s):
Burkholderia cepacia (Taxon ID: 292)
Expression System(s):
Method Details:


