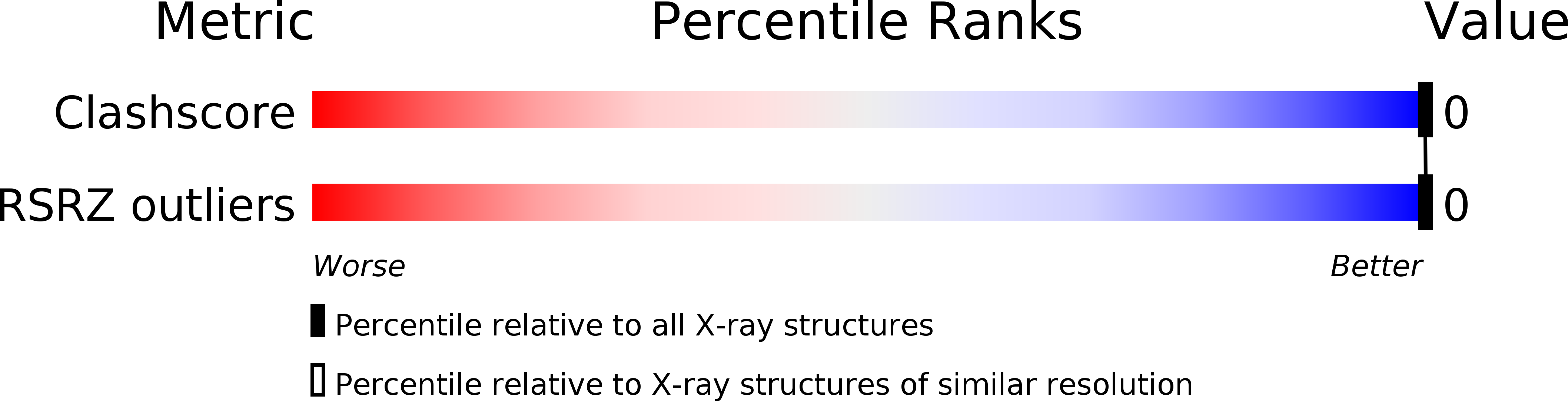
Deposition Date
1992-01-06
Release Date
1993-04-15
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1D54
Keywords:
Title:
ANTHRACYCLINE BINDING TO DNA: HIGH RESOLUTION STRUCTURE OF D(TGTACA) COMPLEXED WITH 4'-EPIADRIAMYCIN
Method Details:
Experimental Method:
Resolution:
1.40 Å
R-Value Observed:
0.17
Space Group:
P 41 21 2