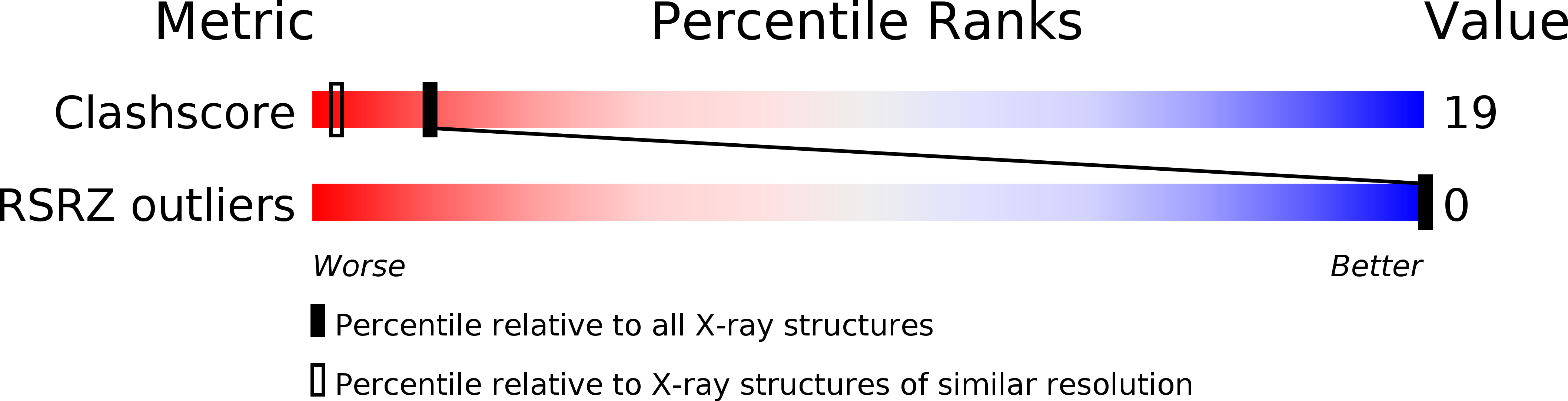
Deposition Date
1991-05-07
Release Date
1992-04-15
Last Version Date
2024-04-03
Entry Detail
PDB ID:
1D41
Keywords:
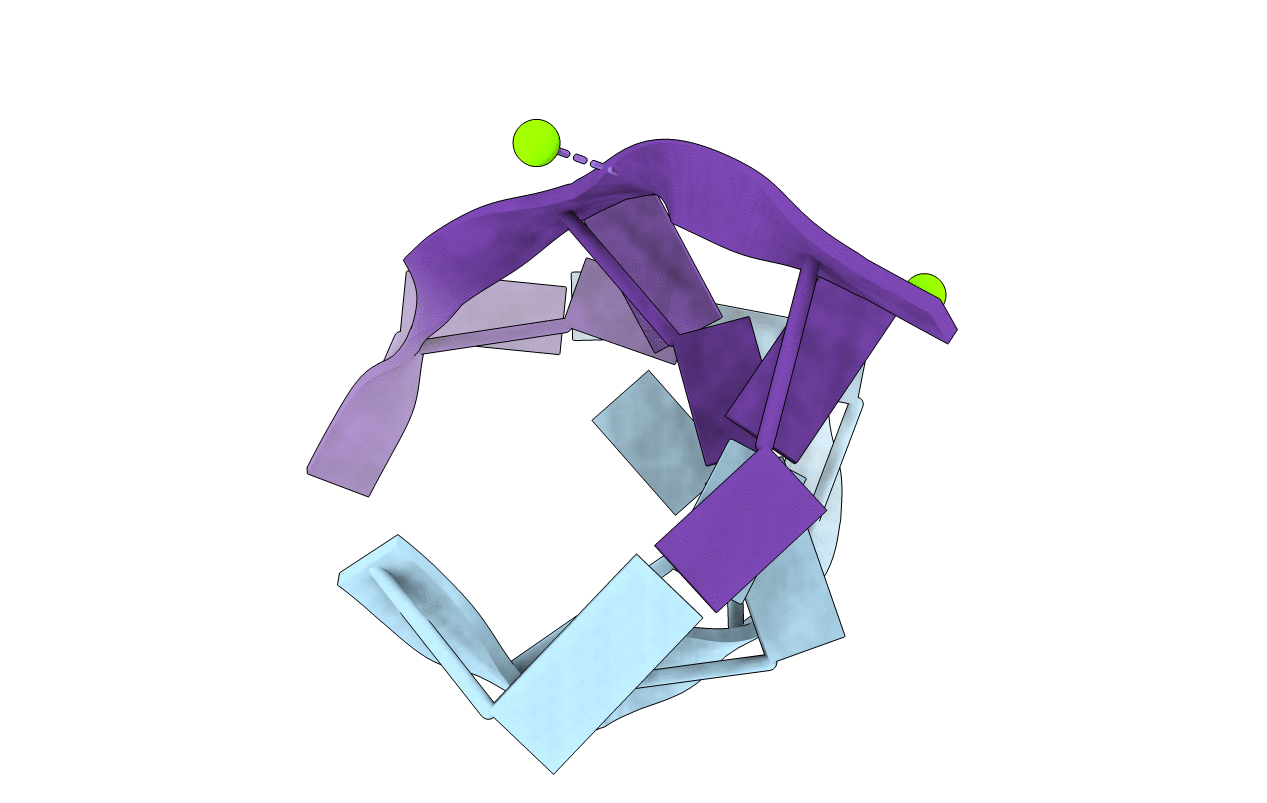
Title:
STABILIZATION OF Z-DNA BY DEMETHYLATION OF THYMINE BASES: 1.3 ANGSTROMS SINGLE-CRYSTAL STRUCTURE OF D(M5CGUAM5CG)
Method Details:
Experimental Method:
Resolution:
1.30 Å
R-Value Observed:
0.20
Space Group:
P 21 21 21