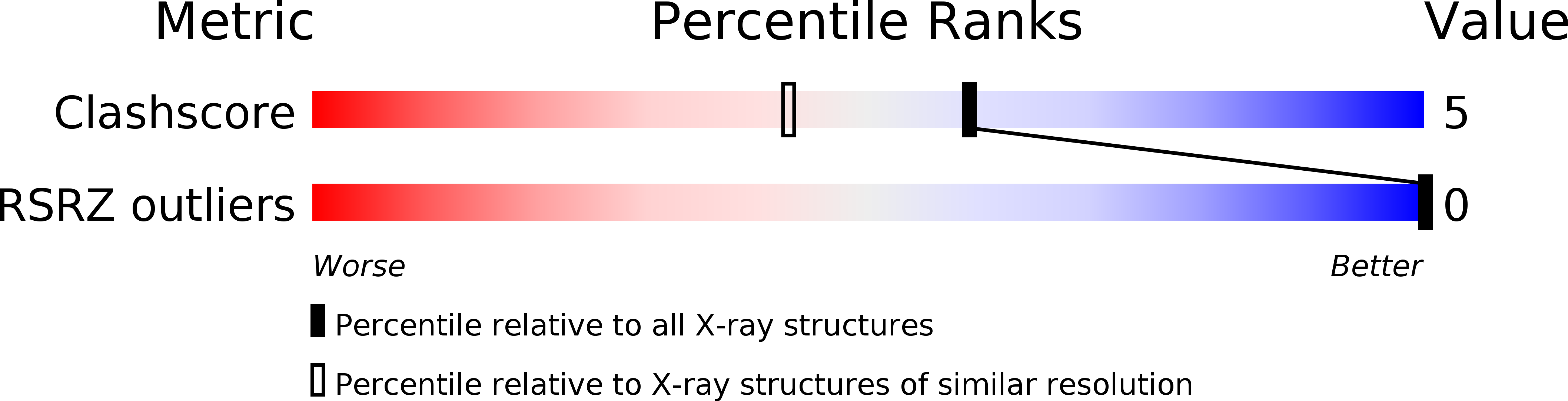Deposition Date
1991-05-29
Release Date
1991-10-15
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1D23
Keywords:
Title:
THE STRUCTURE OF B-HELICAL C-G-A-T-C-G-A-T-C-G AND COMPARISON WITH C-C-A-A-C-G-T-T-G-G. THE EFFECT OF BASE PAIR REVERSALS
Method Details:
Experimental Method:
Resolution:
1.50 Å
R-Value Observed:
0.16
Space Group:
P 21 21 21