Deposition Date
1999-09-21
Release Date
2000-04-02
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1D1U
Keywords:
Title:
USE OF AN N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE TO FACILITATE CRYSTALLIZATION AND ANALYSIS OF A PSEUDO-16-MER DNA MOLECULE CONTAINING G-A MISPAIRS
Biological Source:
Source Organism(s):
Moloney murine leukemia virus (Taxon ID: 11801)
Expression System(s):
Method Details:
Experimental Method:
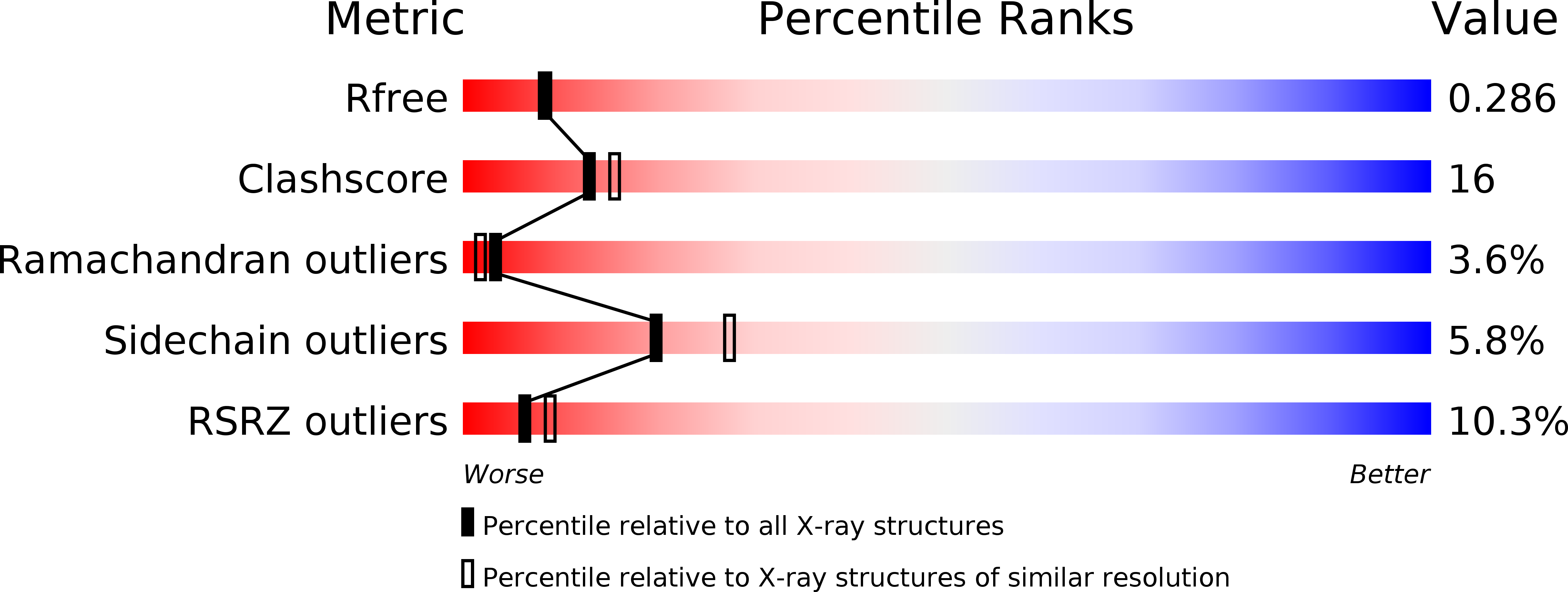
Resolution:
2.30 Å
R-Value Free:
0.28
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 21 21 2