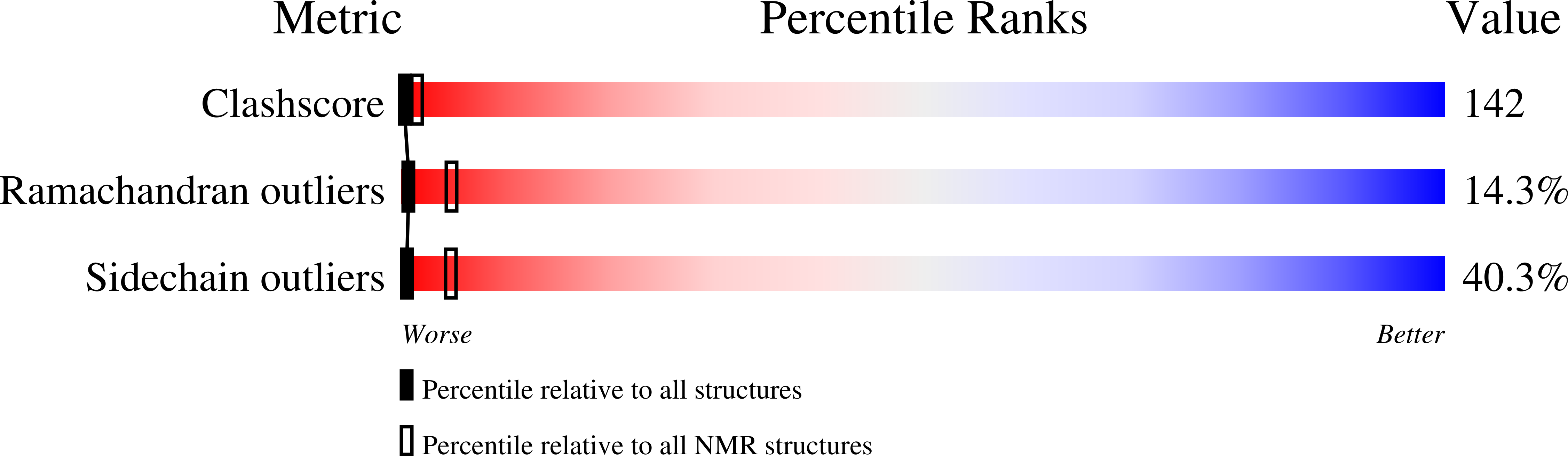
Deposition Date
1995-08-24
Release Date
1995-12-07
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1CYU
Keywords:
Title:
SOLUTION NMR STRUCTURE OF RECOMBINANT HUMAN CYSTATIN A UNDER THE CONDITION OF PH 3.8 AND 310K
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Conformers Submitted:
15