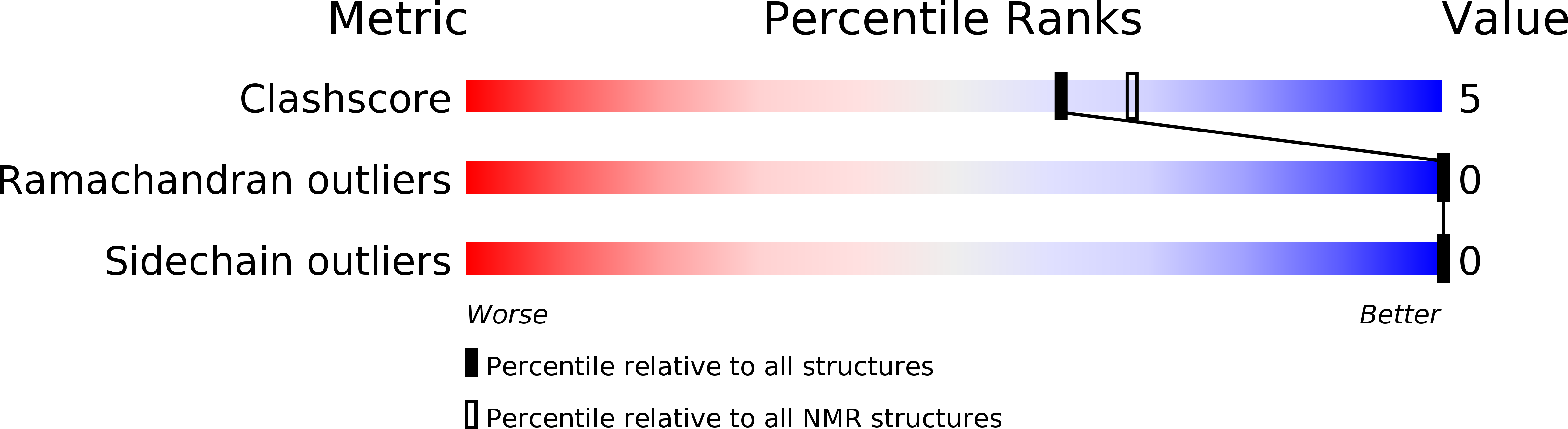
Deposition Date
1992-02-24
Release Date
1994-04-30
Last Version Date
2024-06-05
Entry Detail
PDB ID:
1CSA
Keywords:
Title:
THE MUTANT E.COLI F112W CYCLOPHILIN BINDS CYCLOSPORIN A IN NEARLY IDENTICAL CONFORMATION AS HUMAN CYCLOPHILIN
Biological Source:
Source Organism:
TOLYPOCLADIUM INFLATUM (Taxon ID: 29910)
Method Details:
Experimental Method:
Conformers Submitted:
18