Deposition Date
1994-03-11
Release Date
1994-06-22
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1CPN
Keywords:
Title:
NATIVE-LIKE IN VIVO FOLDING OF A CIRCULARLY PERMUTED JELLYROLL PROTEIN SHOWN BY CRYSTAL STRUCTURE ANALYSIS
Biological Source:
Source Organism(s):
Paenibacillus macerans (Taxon ID: 44252)
Method Details:
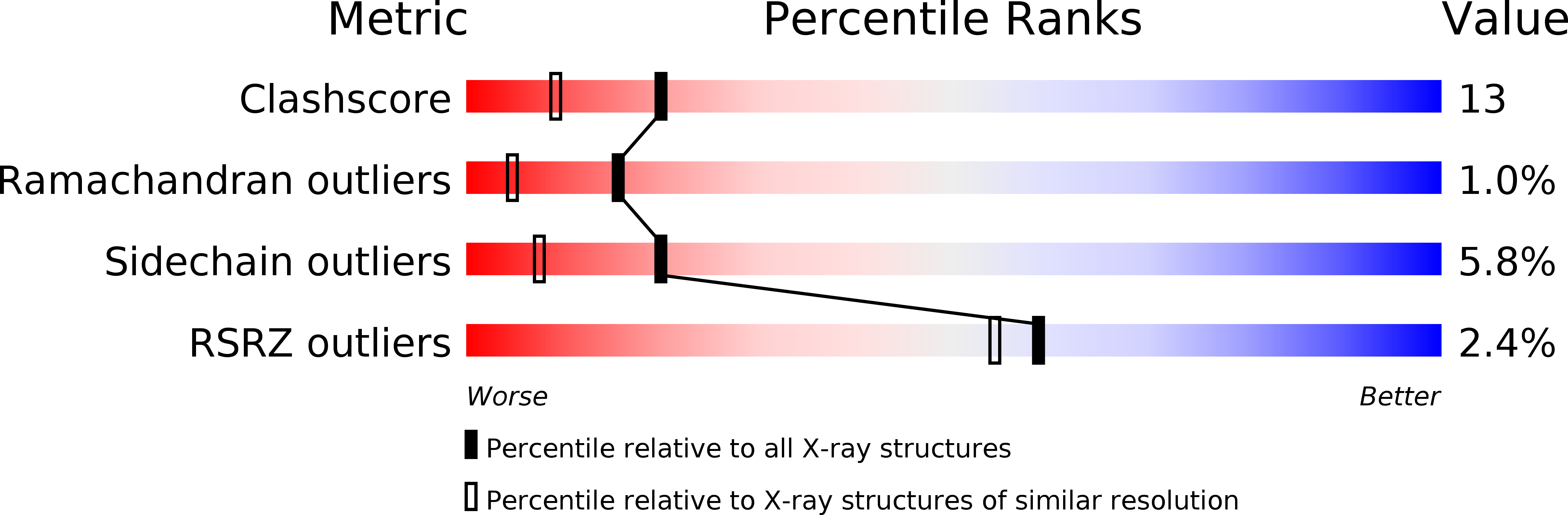
Experimental Method:
Resolution:
1.80 Å
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 1 21 1