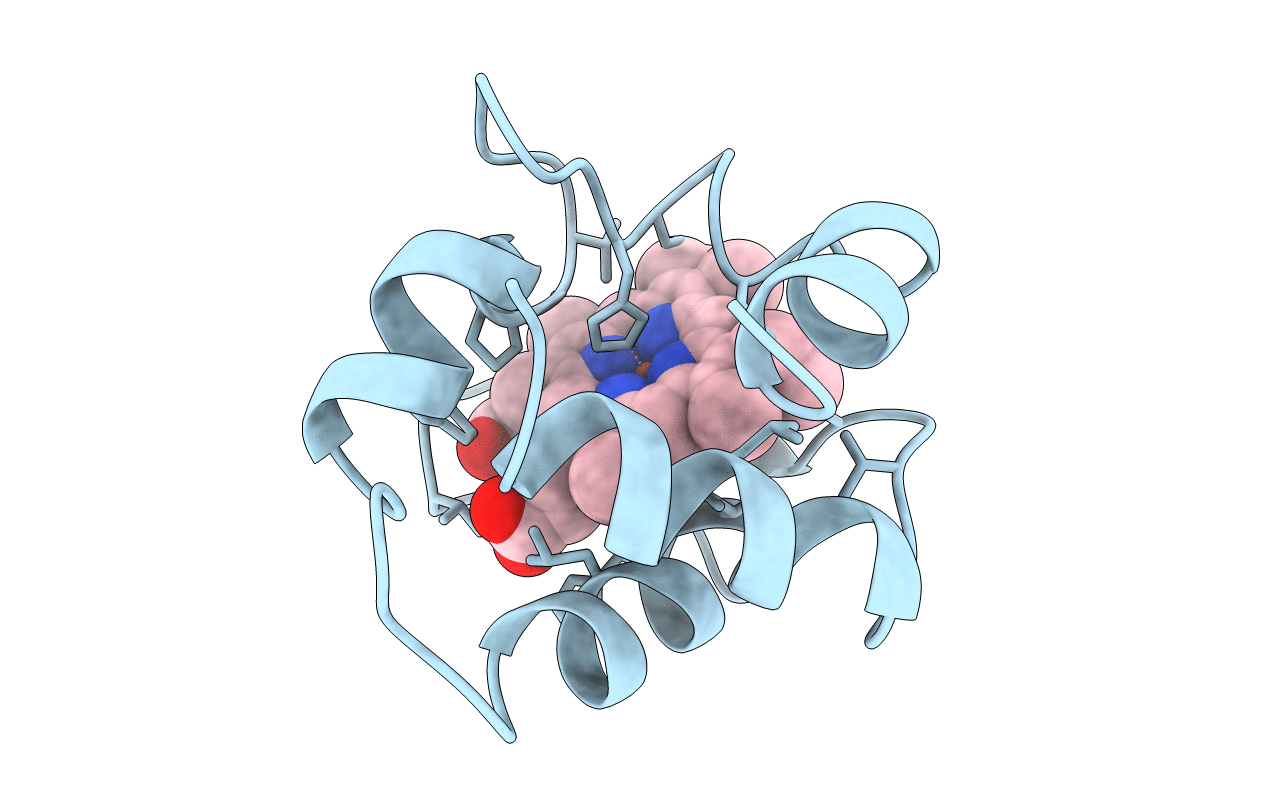
Deposition Date
1993-06-23
Release Date
1993-10-31
Last Version Date
2024-10-30
Entry Detail
PDB ID:
1COR
Keywords:
Title:
INVESTIGATION OF THE SOLUTION CONFORMATION OF CYTOCHROME C-551 FROM PSEUDOMONAS STUTZERI
Biological Source:
Source Organism(s):
Pseudomonas stutzeri (Taxon ID: 316)
Method Details:
Experimental Method:
Conformers Submitted:
1