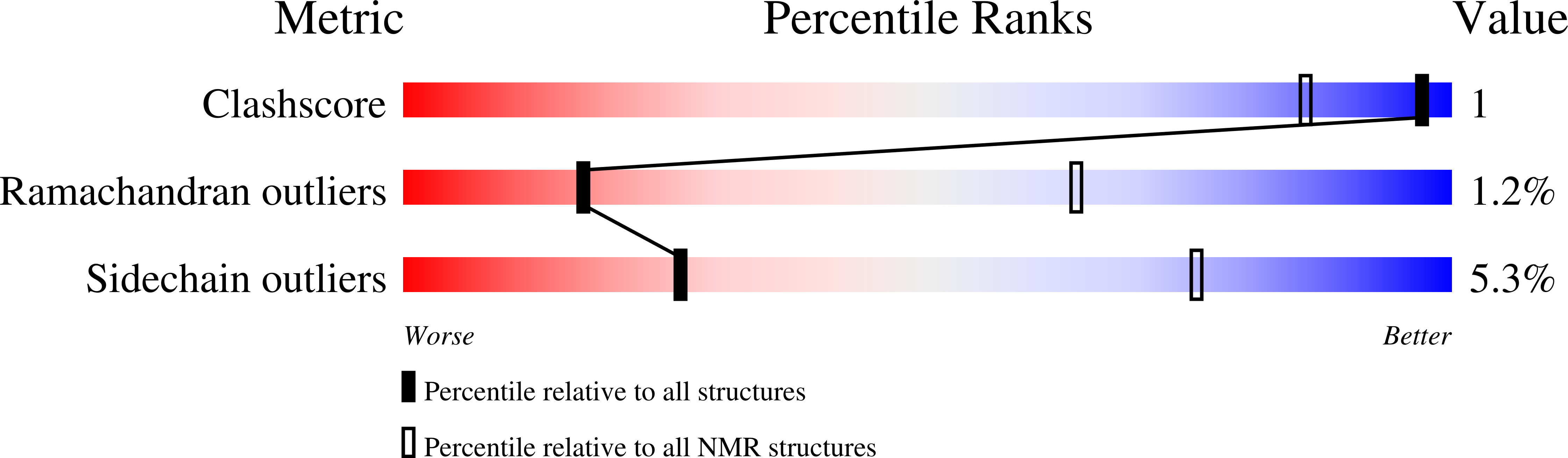
Deposition Date
1995-02-08
Release Date
1995-04-20
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1CLB
Keywords:
Title:
Determination of the solution structure of apo calbindin D9K by nmr spectroscopy
Biological Source:
Source Organism(s):
Bos taurus (Taxon ID: 9913)
Expression System(s):
Method Details:
Experimental Method:
Conformers Submitted:
33