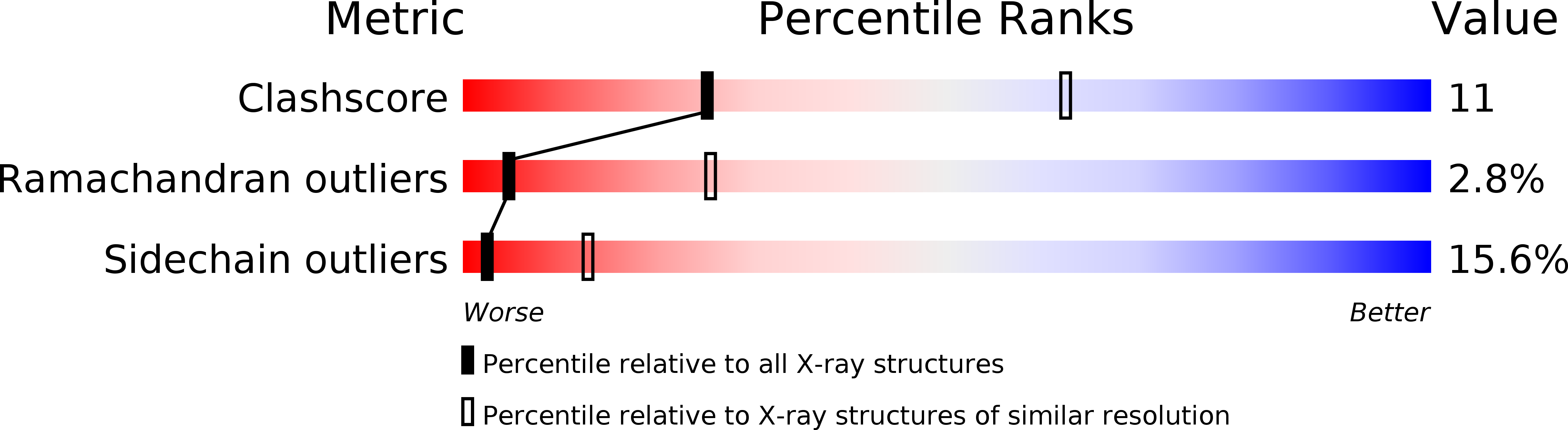
Deposition Date
1991-08-12
Release Date
1994-01-31
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1CGP
Keywords:
Title:
CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Escherichia coli (Taxon ID: 562)
Escherichia coli (Taxon ID: 562)
Method Details: