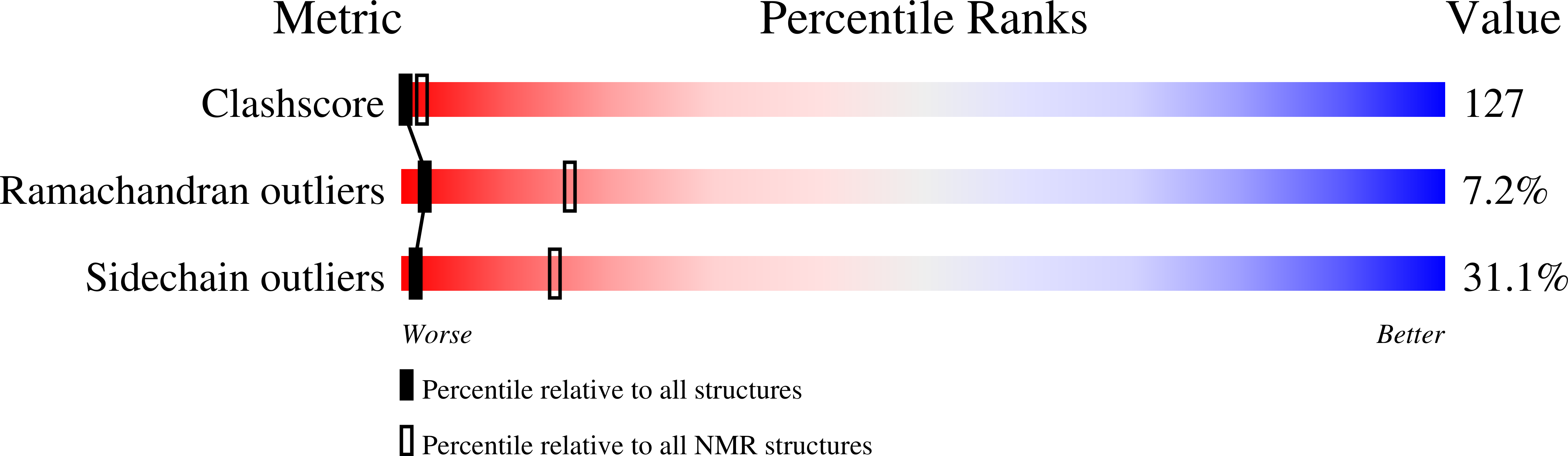
Deposition Date
1996-06-04
Release Date
1997-03-12
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1CFP
Keywords:
Title:
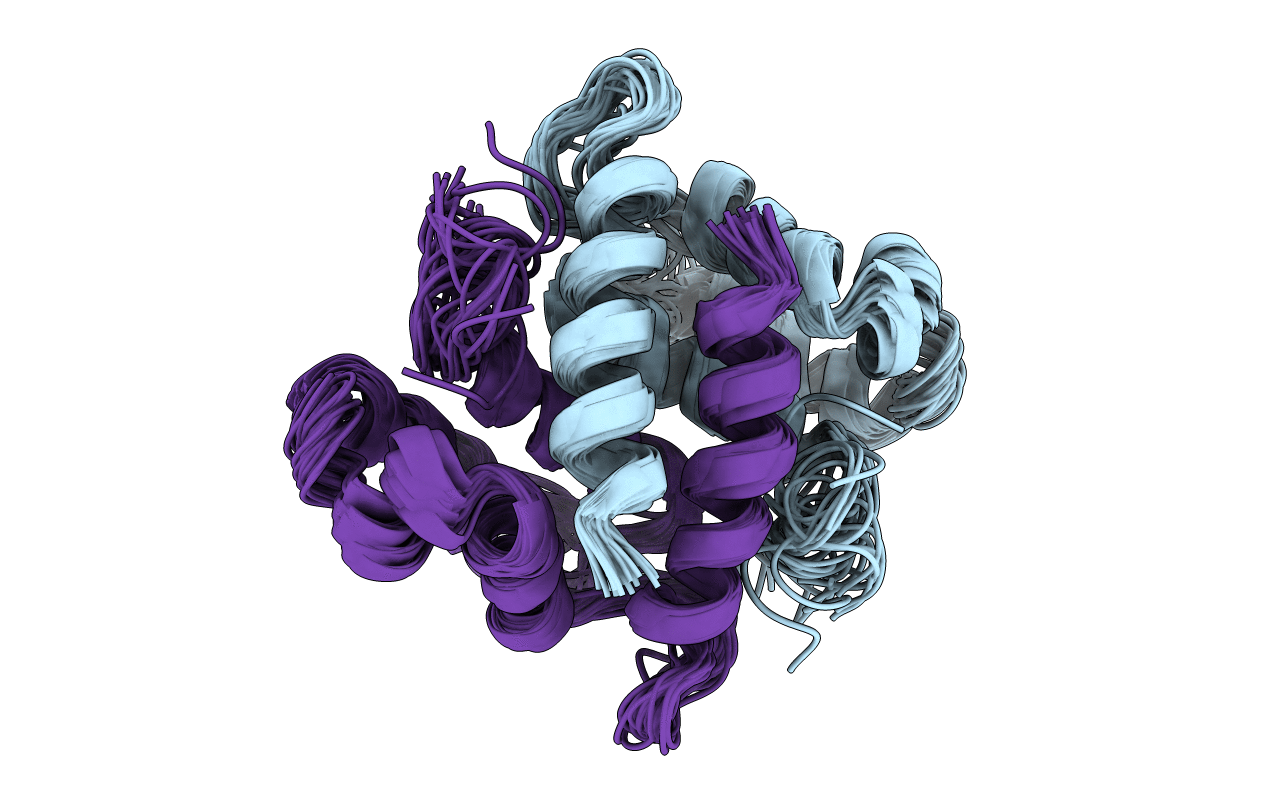
S100B (S100BETA) NMR DATA WAS COLLECTED FROM A SAMPLE OF CALCIUM FREE PROTEIN AT PH 6.3 AND A TEMPERATURE OF 311 K AND 1.7-6.9 MM CONCENTRATION, 25 STRUCTURES
Biological Source:
Source Organism(s):
Bos taurus (Taxon ID: 9913)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
91
Conformers Submitted:
25
Selection Criteria:
TOTAL ENERGY