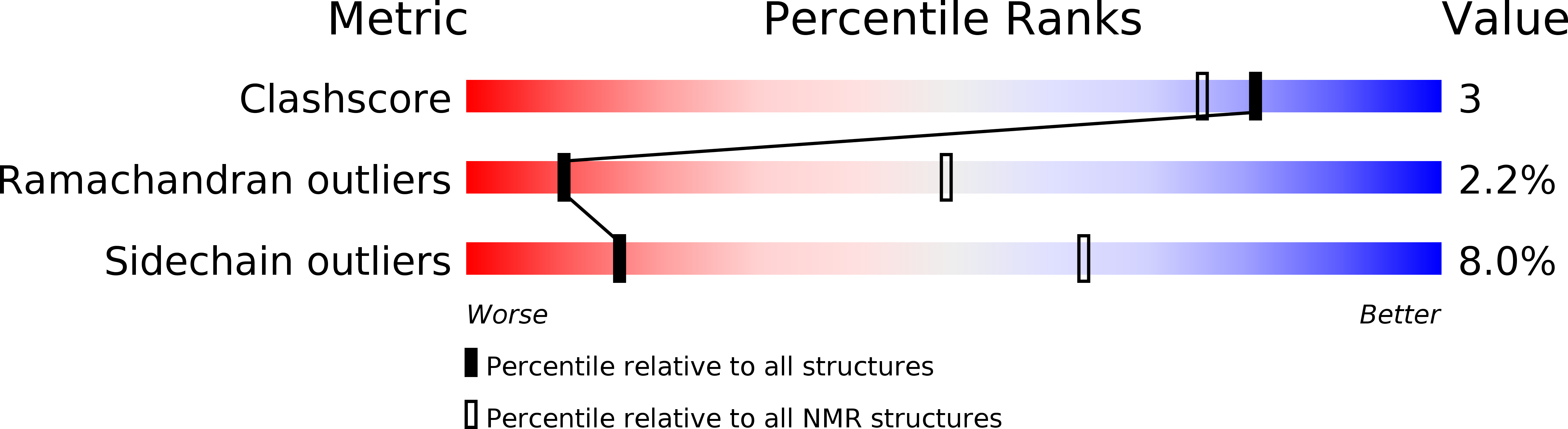
Deposition Date
1993-04-14
Release Date
1993-10-31
Last Version Date
2024-11-20
Entry Detail
PDB ID:
1CCM
Keywords:
Title:
DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL
Biological Source:
Source Organism(s):
Crambe hispanica subsp. abyssinica (Taxon ID: 3721)
Method Details:
Experimental Method:
Conformers Submitted:
8