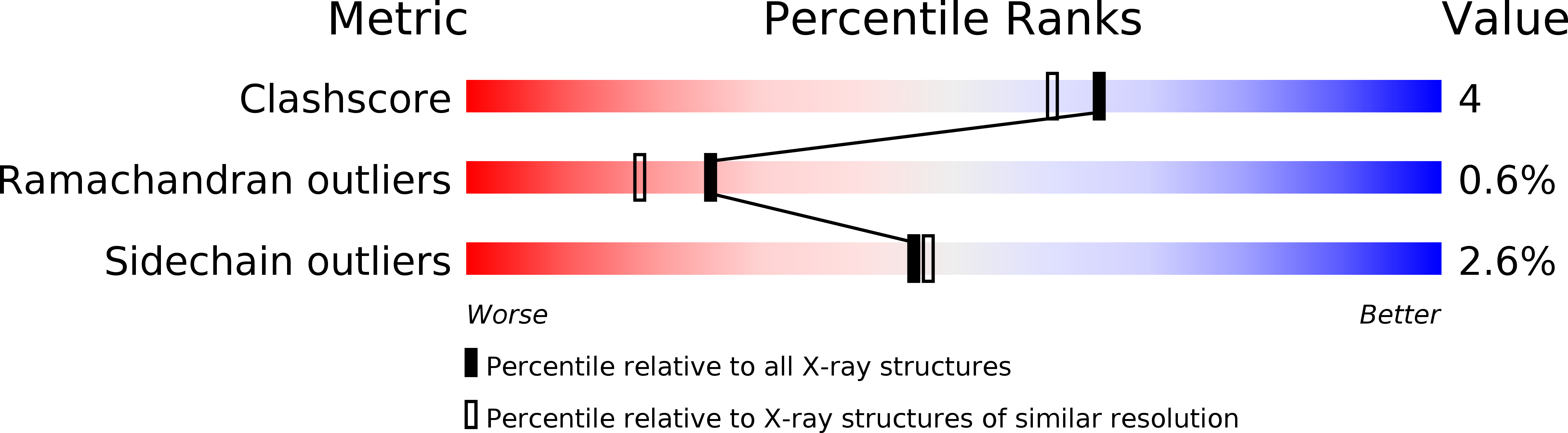Deposition Date
1995-07-31
Release Date
1995-10-15
Last Version Date
2024-10-23
Entry Detail
PDB ID:
1CBG
Keywords:
Title:
THE CRYSTAL STRUCTURE OF A CYANOGENIC BETA-GLUCOSIDASE FROM WHITE CLOVER (TRIFOLIUM REPENS L.), A FAMILY 1 GLYCOSYL-HYDROLASE
Biological Source:
Source Organism(s):
Trifolium repens (Taxon ID: 3899)
Method Details:
Experimental Method:
Resolution:
2.15 Å
R-Value Free:
0.24
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 41 21 2