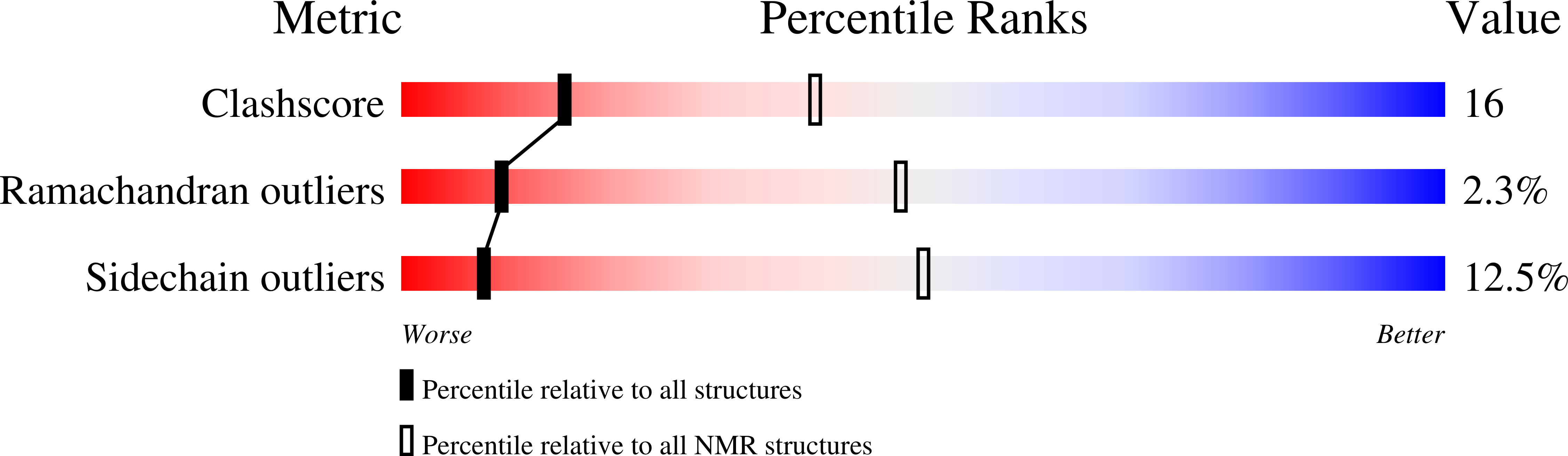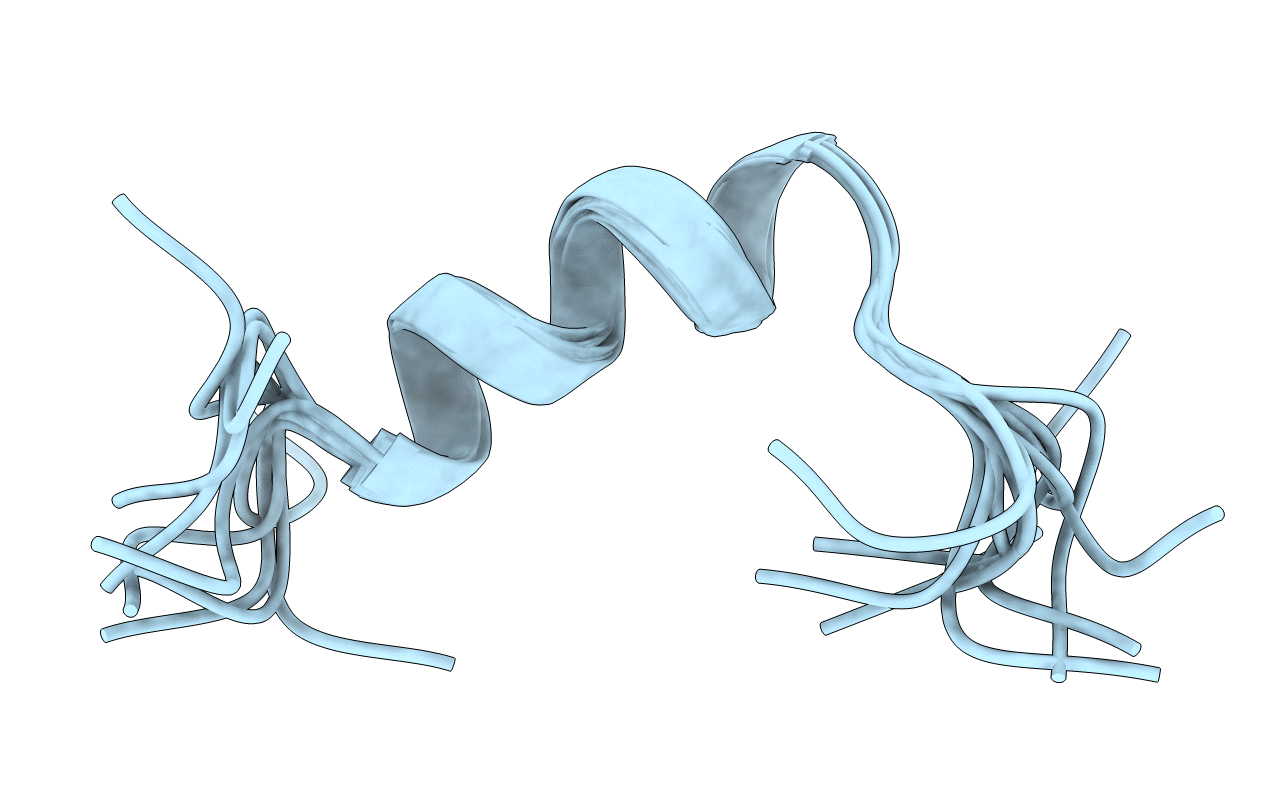
Deposition Date
1998-10-21
Release Date
1999-10-29
Last Version Date
2023-12-27
Entry Detail
Biological Source:
Source Organism(s):
Rattus norvegicus (Taxon ID: 10116)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
25
Conformers Submitted:
10
Selection Criteria:
NO VIOLATIONS >0.3 DEGREES OR 3 ANGSTROMS; LOWEST ENERGY