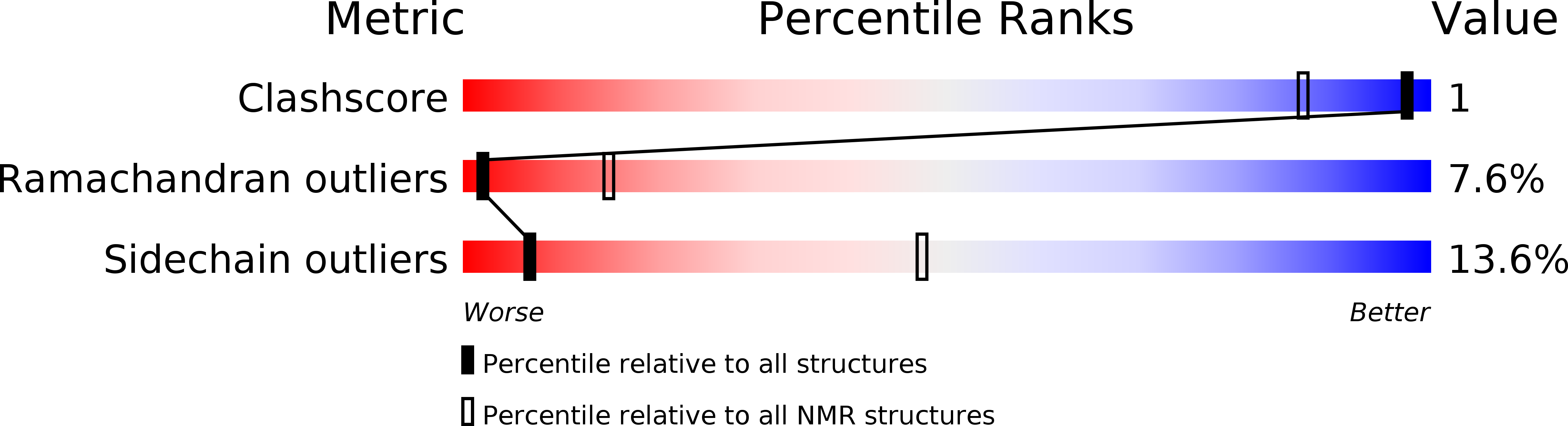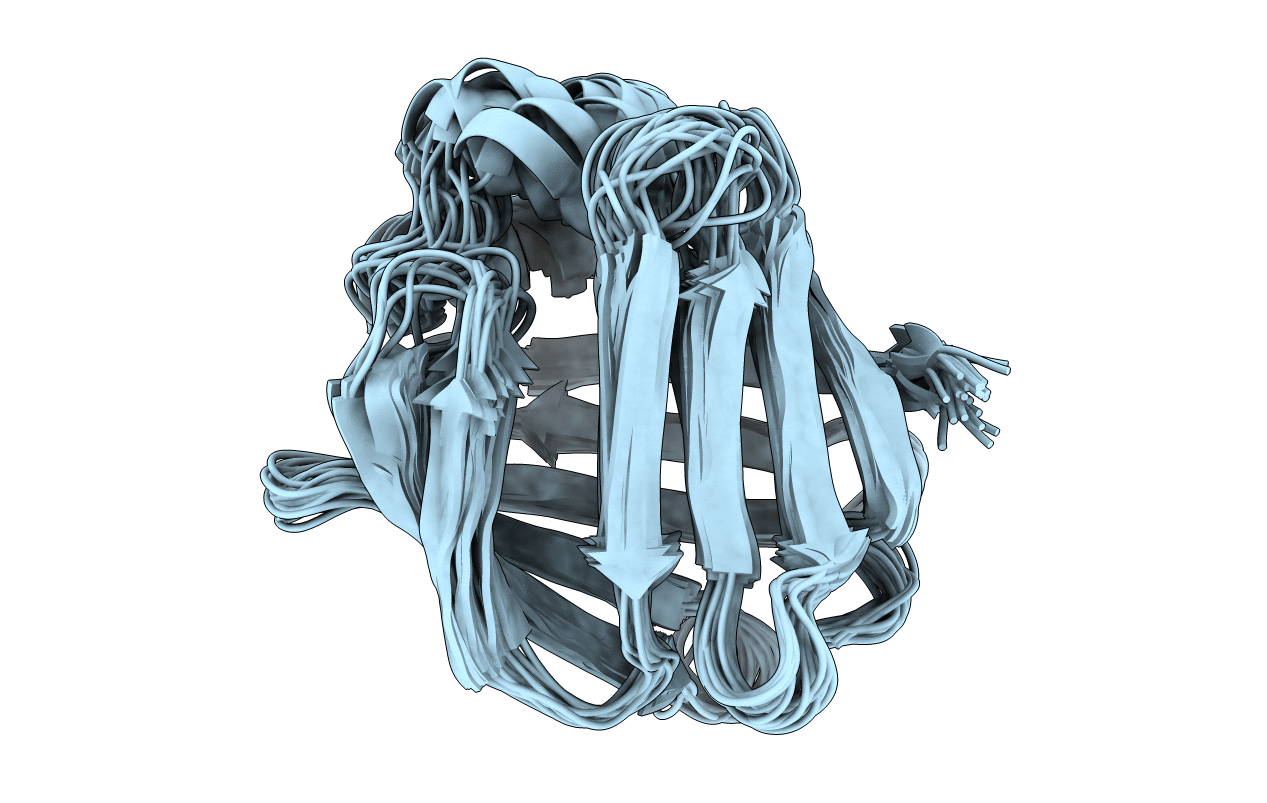
Deposition Date
1998-09-29
Release Date
1998-10-07
Last Version Date
2024-04-10
Entry Detail
PDB ID:
1BWY
Keywords:
Title:
NMR STUDY OF BOVINE HEART FATTY ACID BINDING PROTEIN
Biological Source:
Source Organism(s):
Bos taurus (Taxon ID: 9913)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
25
Selection Criteria:
LOWEST VIOLATION OF EXPERIMENTAL DISTANCE CONSTRAINTS