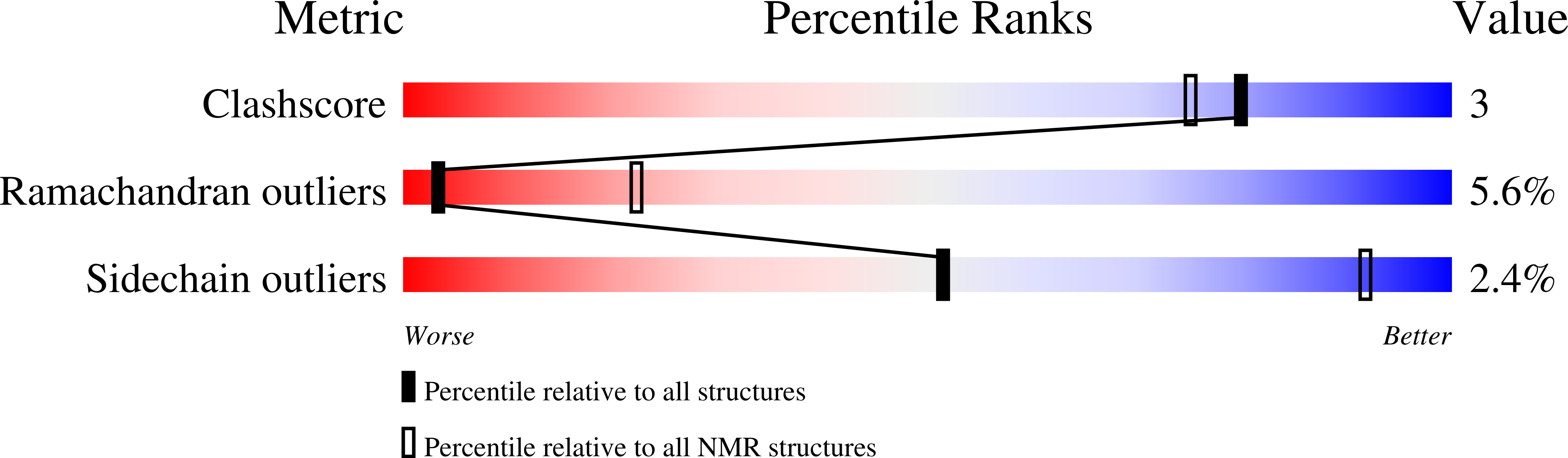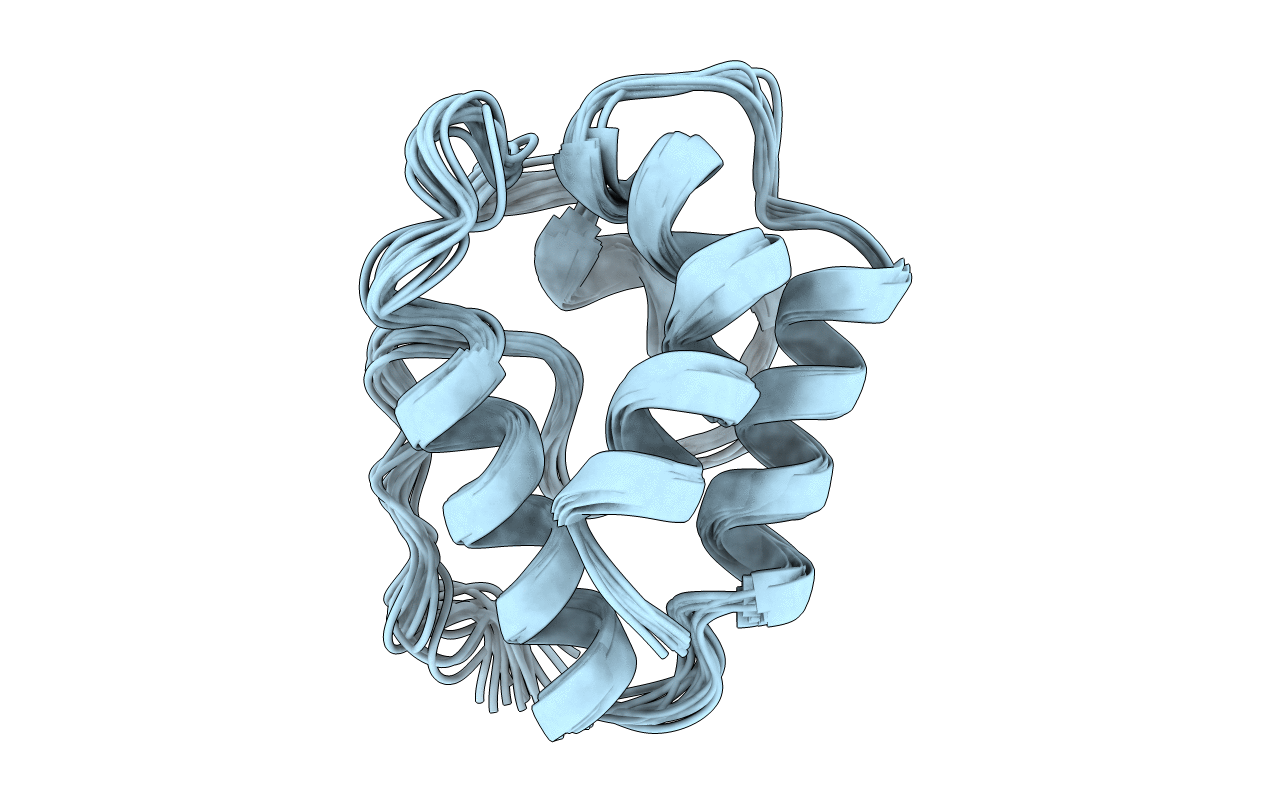
Deposition Date
1998-09-21
Release Date
1999-05-18
Last Version Date
2024-11-20
Entry Detail
PDB ID:
1BV2
Keywords:
Title:
LIPID TRANSFER PROTEIN FROM RICE SEEDS, NMR, 14 STRUCTURES
Biological Source:
Source Organism(s):
Oryza sativa (Taxon ID: 4530)
Method Details:
Experimental Method:
Conformers Calculated:
200
Conformers Submitted:
14
Selection Criteria:
LEAST RESTRAINT VIOLATION AND ANALYSIS OF THE RAMACHANDRAN MAPS AND ENERGETIC CRITERIA