Deposition Date
1998-08-30
Release Date
1998-09-16
Last Version Date
2023-08-09
Entry Detail
PDB ID:
1BU6
Keywords:
Title:
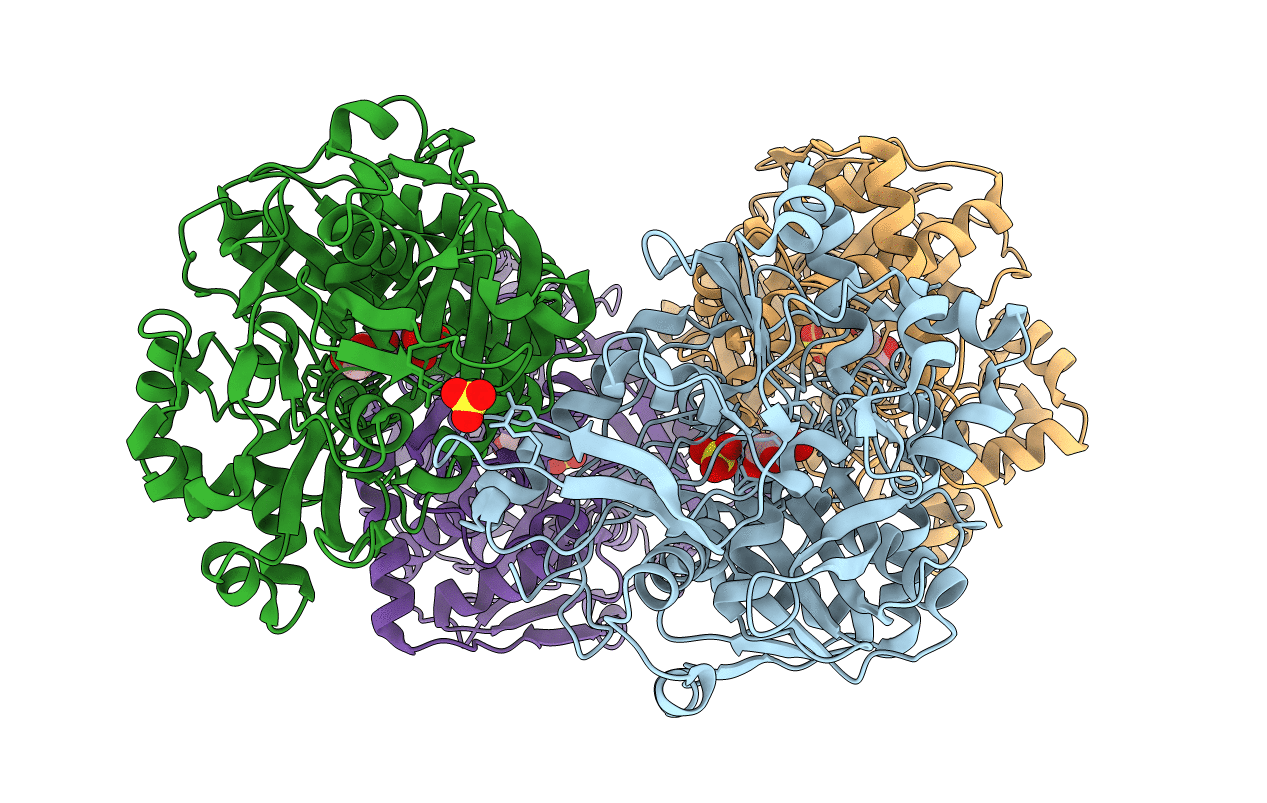
CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details: