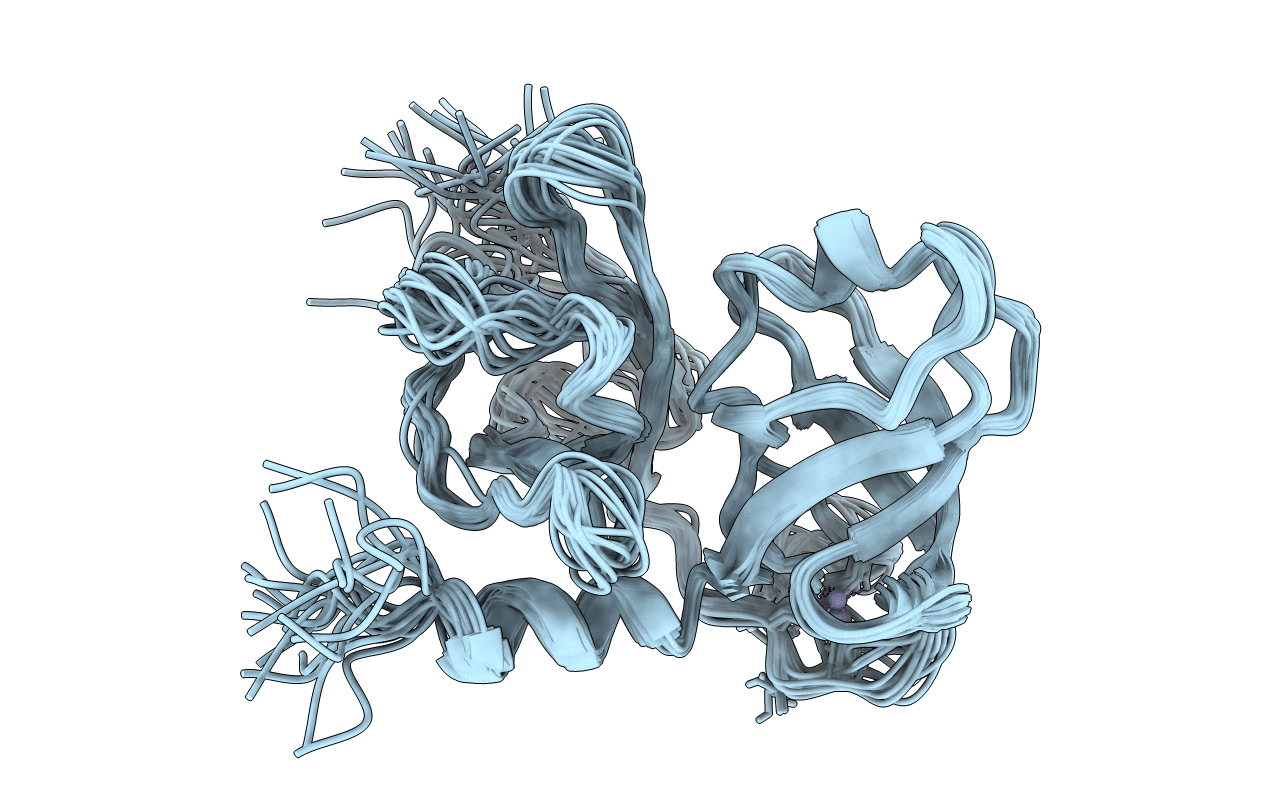
Deposition Date
1998-09-01
Release Date
1999-06-22
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1BT7
Keywords:
Title:
THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES
Biological Source:
Source Organism(s):
Hepatitis C virus (Taxon ID: 11103)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
60
Conformers Submitted:
20
Selection Criteria:
LEAST RESTRAINT VIOLATIONS/MINIMUM ENERGY


