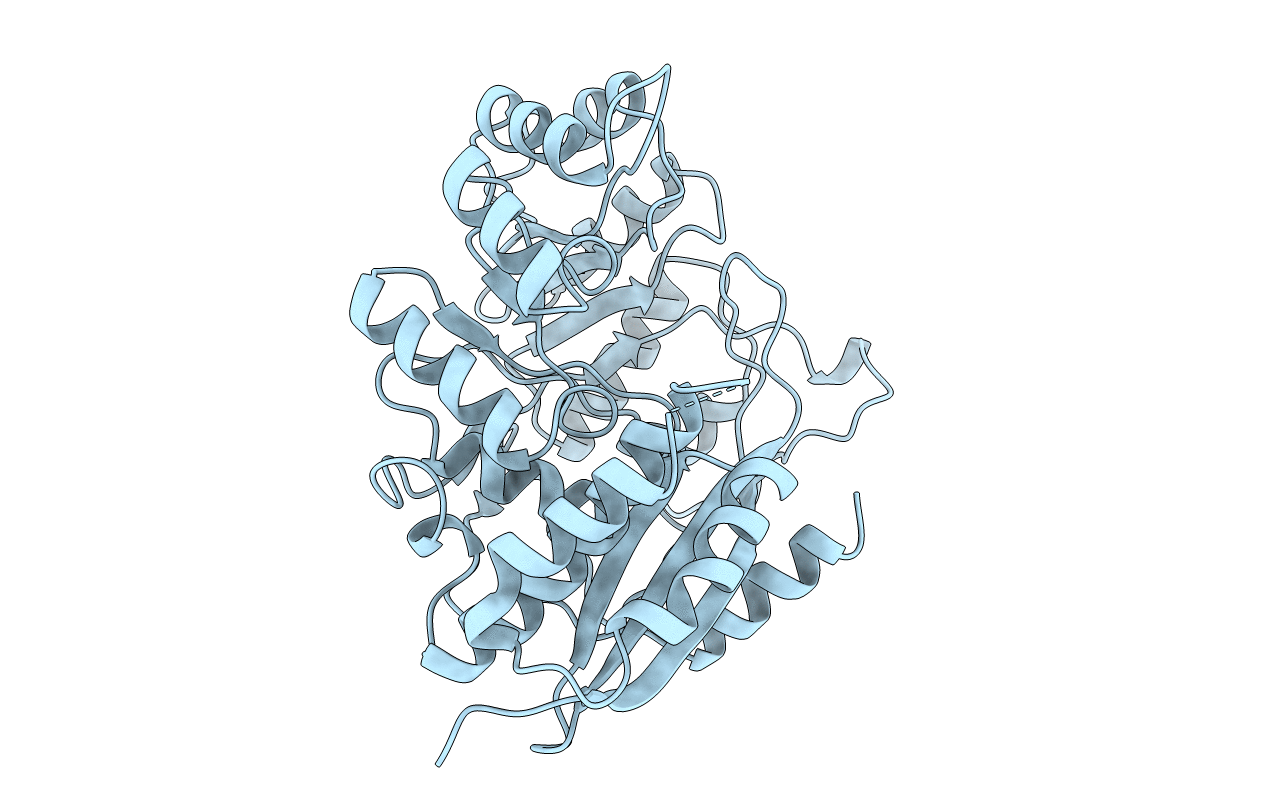Abstact
The structure of (D)-glucarate dehydratase from Pseudomonas putida (GlucD) has been solved at 2.3 A resolution by multiple isomorphous replacement and refined to a final R-factor of 19.0%. The protein crystallizes in the space group I222 with one subunit in the asymmetric unit. The unit cell dimensions are a = 69.6 A, b = 108.8 A, and c = 122.6 A. The crystals were grown using the batch method where the primary precipitant was poly(ethylene glycol) 1000. The structure reveals that GlucD is a tetramer of four identical polypeptides, each containing 451 residues. The structure was determined without a bound substrate or substrate analogue. Three disordered regions are noted: the N-terminus through residue 11, a loop containing residues 99 through 110, and the C-terminus from residue 423. On the basis of primary sequence alignments, we previously concluded that GlucD is a member of the mandelate racemase (MR) subfamily of the enolase superfamily [Babbitt, P. C., Hasson, M. S., Wedekind, J. E., Palmer, D. R. J., Barrett, W. C., Reed, G. J., Rayment, I., Ringe, D., Kenyon, G. L., and Gerlt, J. A. (1996) Biochemistry 35, 16489-16501]. This prediction is now verified, since the overall fold of GlucD is strikingly similar to those of MR, muconate lactonizing enzyme I, and enolase. Also, many of the active site residues of GlucD can be superimposed on those found in the active site of MR. The implications of this structure on the evolution of catalysis in the enolase superfamily are discussed.