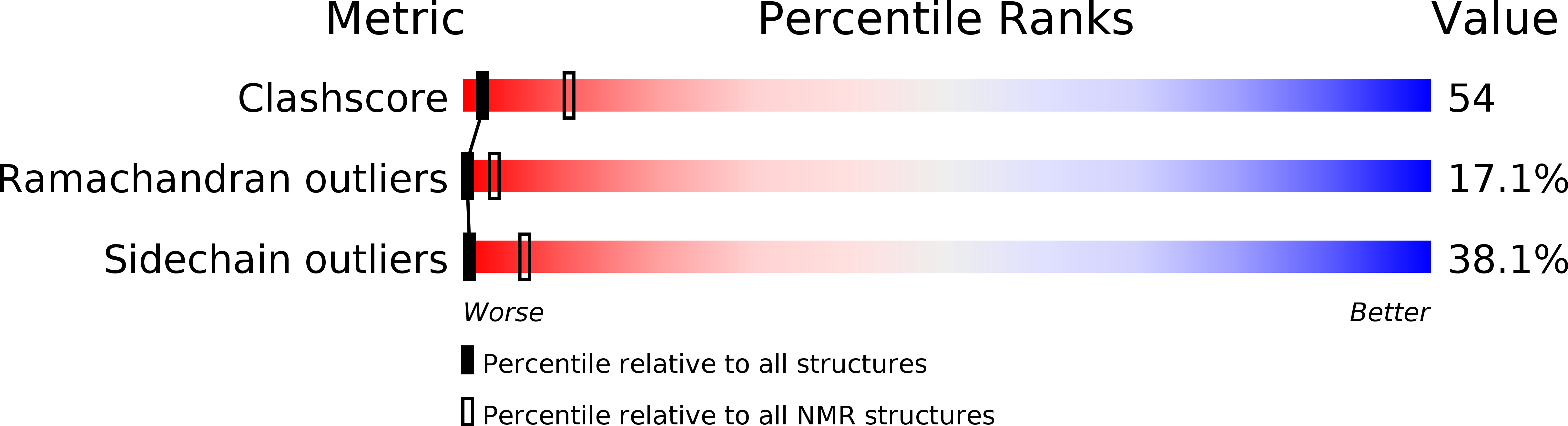Deposition Date
1994-07-21
Release Date
1995-01-26
Last Version Date
2024-10-30
Entry Detail
PDB ID:
1BON
Keywords:
Title:
THREE-DIMENSIONAL STRUCTURE OF BOMBYXIN-II, AN INSULIN-RELATED BRAIN-SECRETORY PEPTIDE OF THE SILKMOTH BOMBYX MORI: COMPARISON WITH INSULIN AND RELAXIN
Biological Source:
Source Organism(s):
Bombyx mori (Taxon ID: 7091)
Method Details:
Experimental Method:
Conformers Submitted:
10