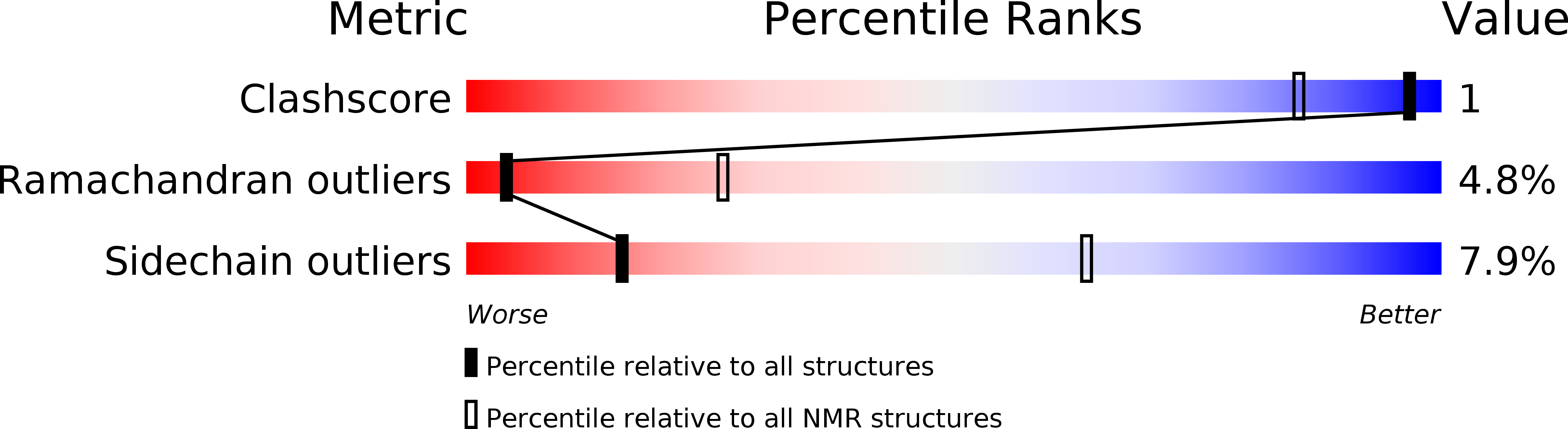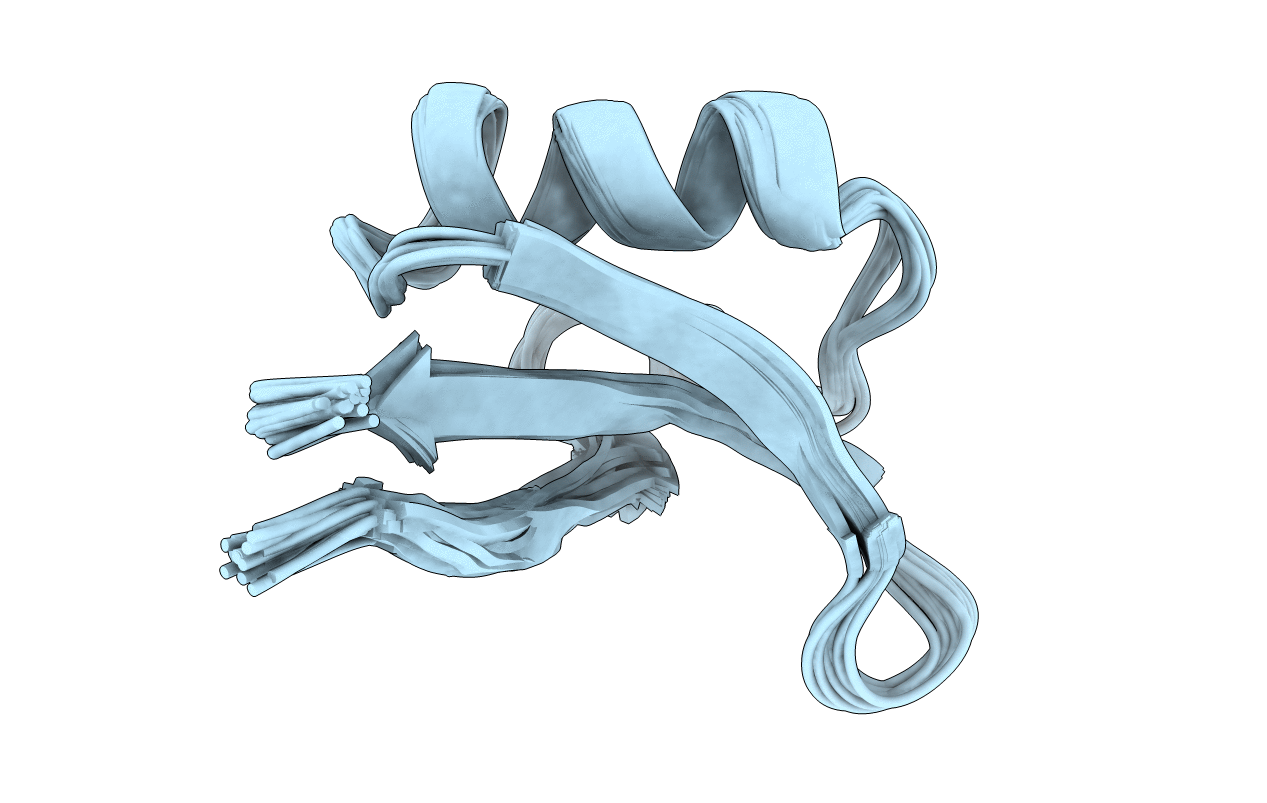
Deposition Date
1998-07-15
Release Date
2000-01-05
Last Version Date
2024-10-30
Entry Detail
PDB ID:
1BK8
Keywords:
Title:
DETERMINATION OF THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF AESCULUS HIPPOCASTANUM ANTIMICROBIAL PROTEIN 1 (AH-AMP1) BY 1H NMR, 25 STRUCTURES
Biological Source:
Source Organism(s):
Aesculus hippocastanum (Taxon ID: 43364)
Method Details:
Experimental Method:
Conformers Calculated:
500
Conformers Submitted:
25
Selection Criteria:
LEAST RESTRAINT VIOLATION AND ENERGY