Deposition Date
1998-06-15
Release Date
1998-11-04
Last Version Date
2024-11-20
Entry Detail
PDB ID:
1BH6
Keywords:
Title:
SUBTILISIN DY IN COMPLEX WITH THE SYNTHETIC INHIBITOR N-BENZYLOXYCARBONYL-ALA-PRO-PHE-CHLOROMETHYL KETONE
Biological Source:
Source Organism(s):
Bacillus licheniformis (Taxon ID: 1402)
Method Details:
Experimental Method:
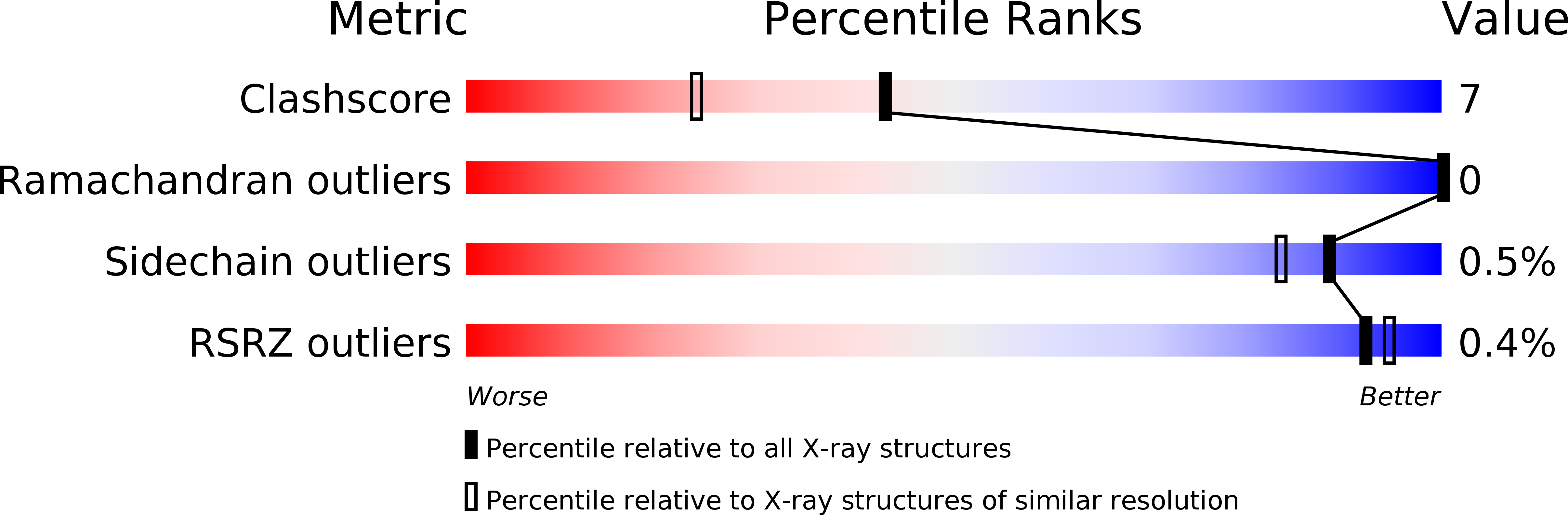
Resolution:
1.75 Å
R-Value Observed:
0.14
Space Group:
P 21 21 21