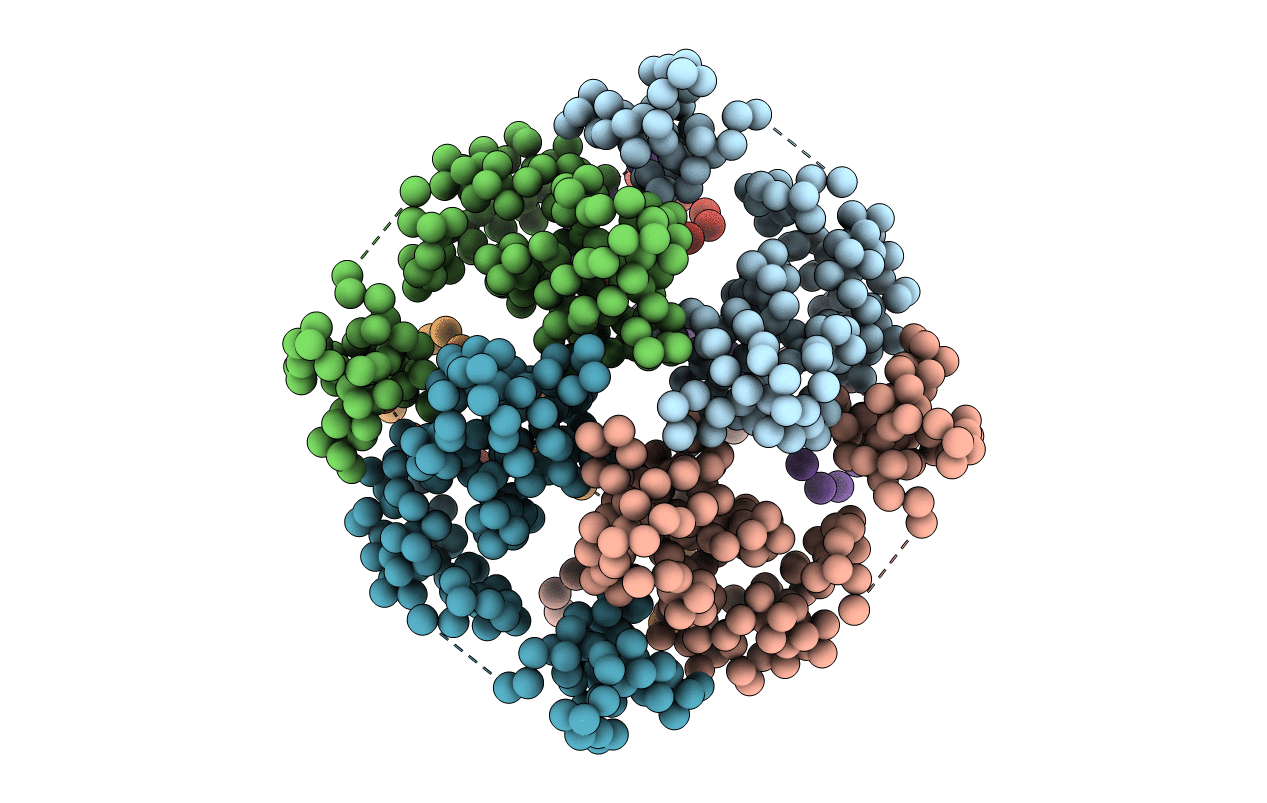
Deposition Date
1998-05-11
Release Date
1999-11-24
Last Version Date
2023-08-09
Entry Detail
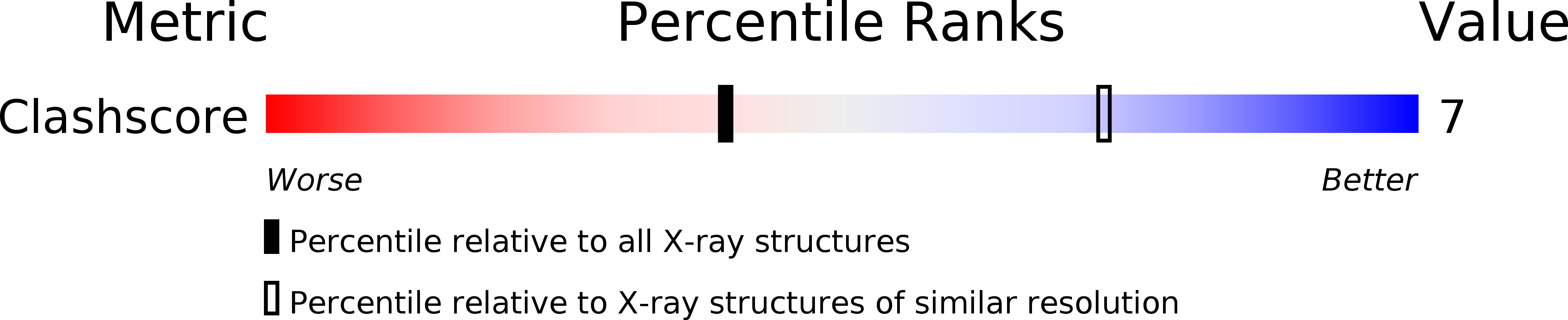
PDB ID:
1BDX
Keywords:
Title:
E. COLI DNA HELICASE RUVA WITH BOUND DNA HOLLIDAY JUNCTION, ALPHA CARBONS AND PHOSPHATE ATOMS ONLY
Biological Source:
Source Organism(s):
Escherichia coli BL21(DE3) (Taxon ID: 469008)
Expression System(s):
Method Details: