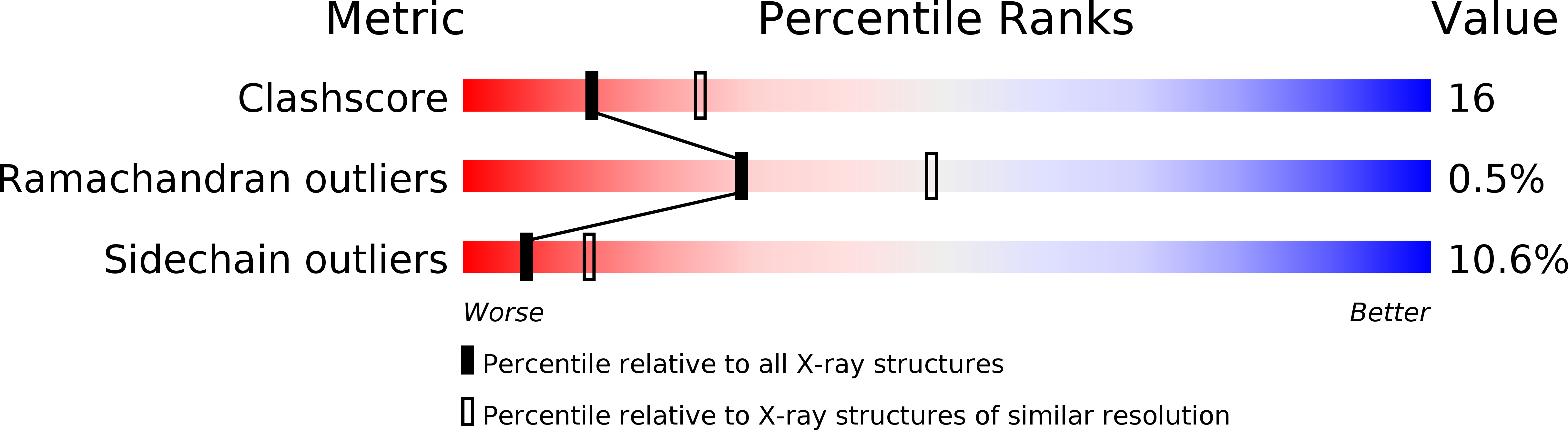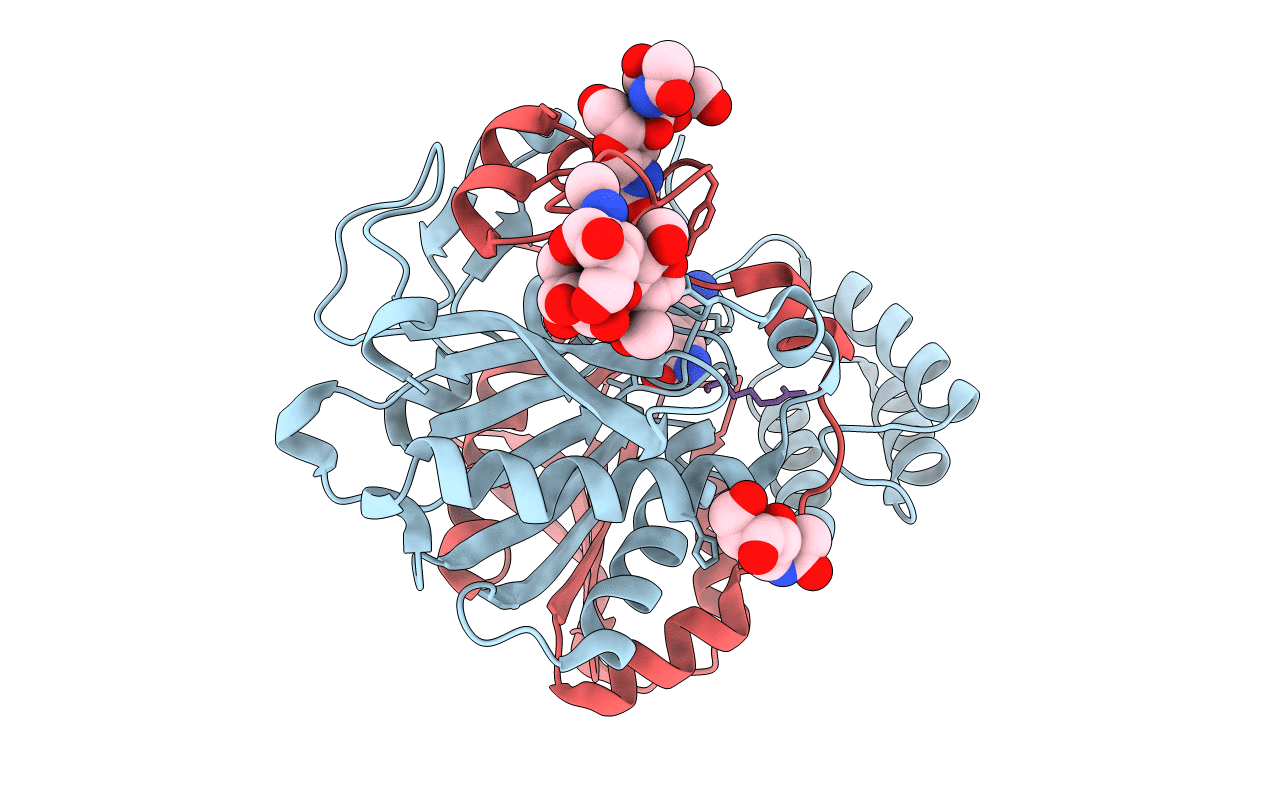
Deposition Date
1995-11-03
Release Date
1996-03-08
Last Version Date
2023-11-15
Entry Detail
PDB ID:
1BCR
Keywords:
Title:
COMPLEX OF THE WHEAT SERINE CARBOXYPEPTIDASE, CPDW-II, WITH THE MICROBIAL PEPTIDE ALDEHYDE INHIBITOR, ANTIPAIN, AND ARGININE AT ROOM TEMPERATURE
Biological Source:
Source Organism(s):
Actinobacteria (Taxon ID: 1760)
Triticum aestivum (Taxon ID: 4565)
Triticum aestivum (Taxon ID: 4565)
Method Details:
Experimental Method:
Resolution:
2.50 Å
R-Value Observed:
0.16
Space Group:
P 41 21 2