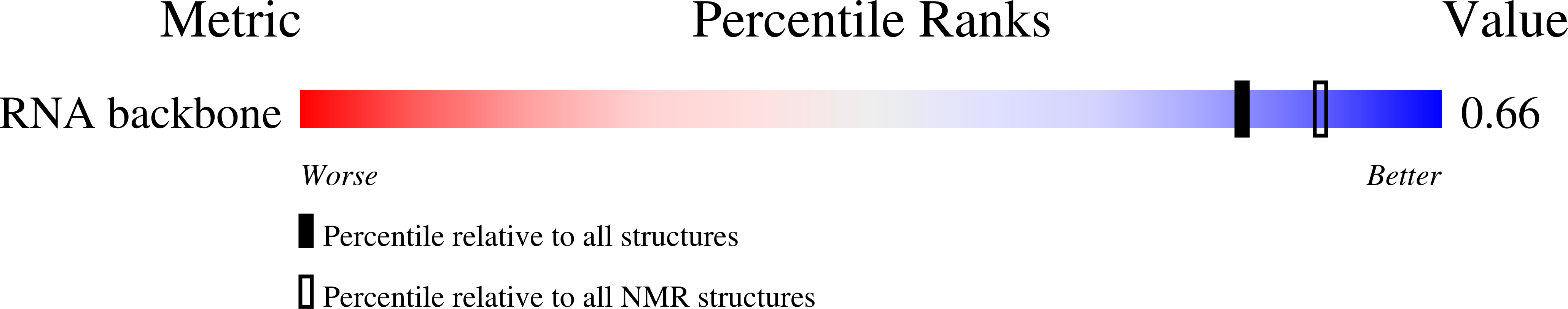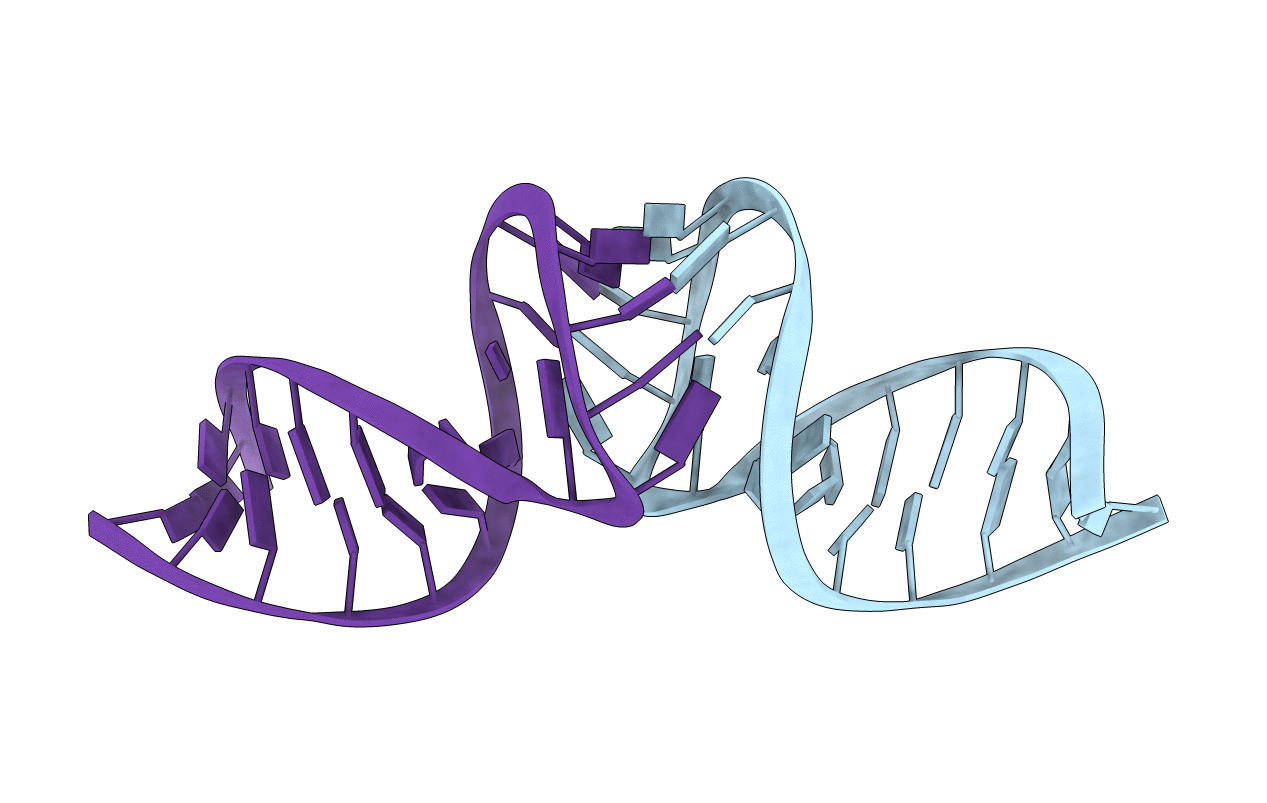
Deposition Date
1998-04-18
Release Date
1999-04-27
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1BAU
Keywords:
Title:
NMR STRUCTURE OF THE DIMER INITIATION COMPLEX OF HIV-1 GENOMIC RNA, MINIMIZED AVERAGE STRUCTURE
Method Details:
Experimental Method:
Conformers Calculated:
34
Conformers Submitted:
1
Selection Criteria:
LOWEST NMR CONSTRAINT VIOLATIONS AND LOWEST AMBER ENERGY.