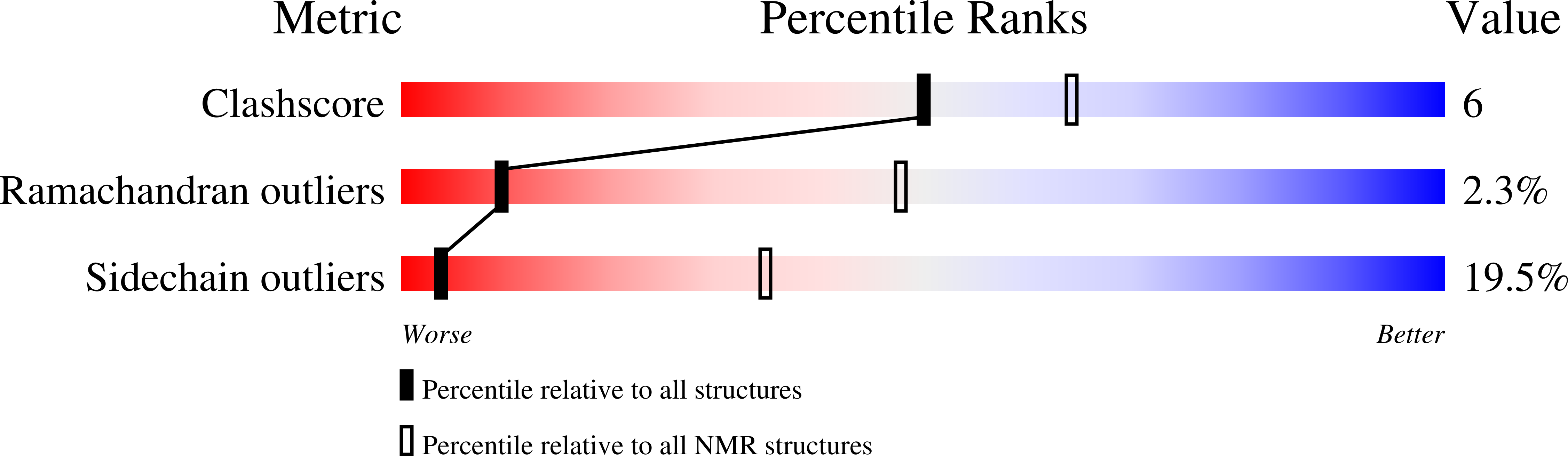
Deposition Date
1998-04-07
Release Date
1998-06-17
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1BA4
Keywords:
Title:
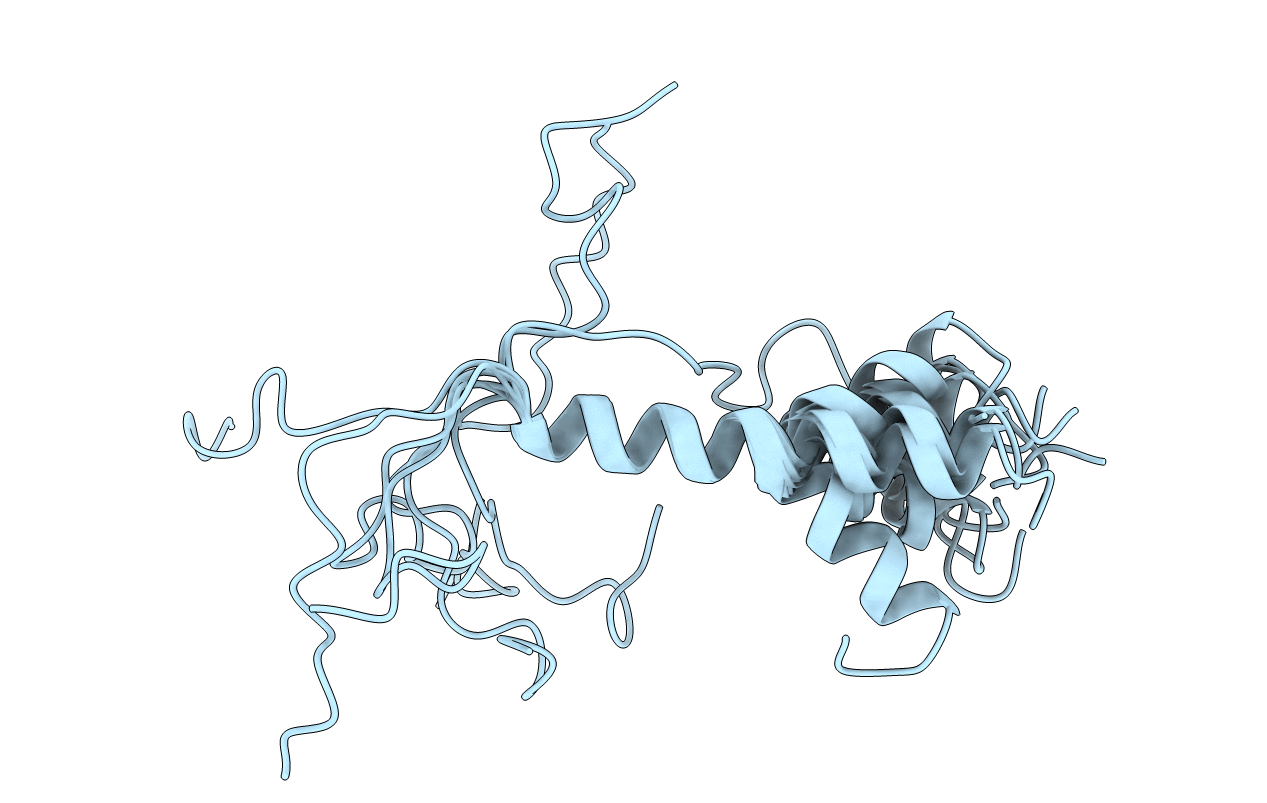
THE SOLUTION STRUCTURE OF AMYLOID BETA-PEPTIDE (1-40) IN A WATER-MICELLE ENVIRONMENT. IS THE MEMBRANE-SPANNING DOMAIN WHERE WE THINK IT IS? NMR, 10 STRUCTURES
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
50
Conformers Submitted:
10
Selection Criteria:
LOW ENERGY, VIOLATIONS AND RMSD