Deposition Date
1998-11-12
Release Date
1999-08-31
Last Version Date
2023-12-27
Entry Detail
PDB ID:
1B0S
Keywords:
Title:
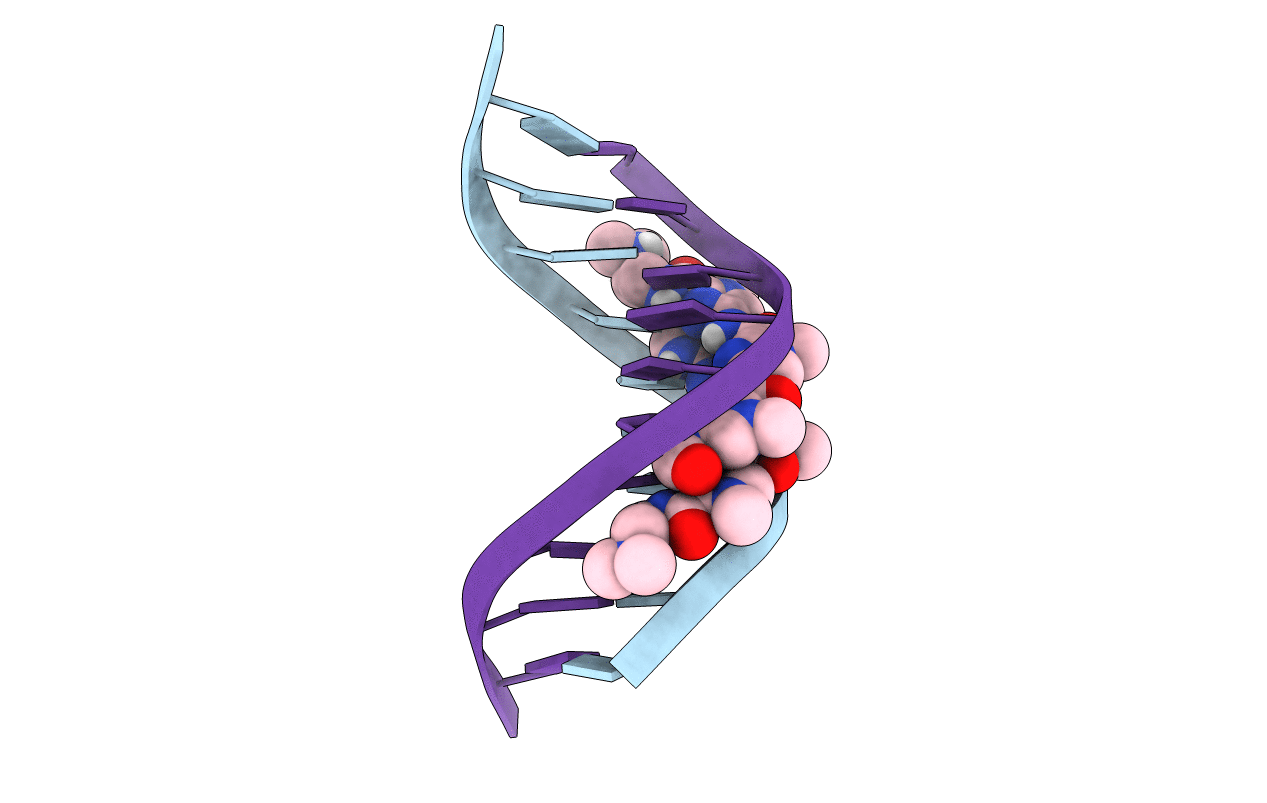
BINDING OF AR-1-144, A TRI-IMIDAZOLE DNA MINOR GROOVE BINDER, TO CCGG SEQUENCE ANALYZED BY NMR SPECTROSCOPY
Method Details:
Experimental Method:
Conformers Calculated:
11
Conformers Submitted:
1
Selection Criteria:
REFINED AVERAGE STRUCTURE