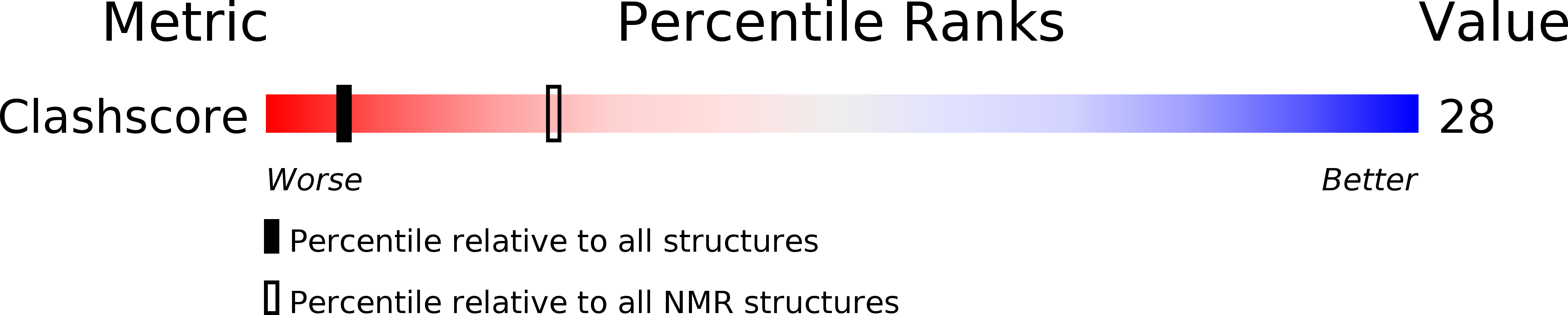
Deposition Date
1997-09-11
Release Date
1998-02-25
Last Version Date
2024-04-10
Entry Detail
PDB ID:
1AU5
Keywords:
Title:
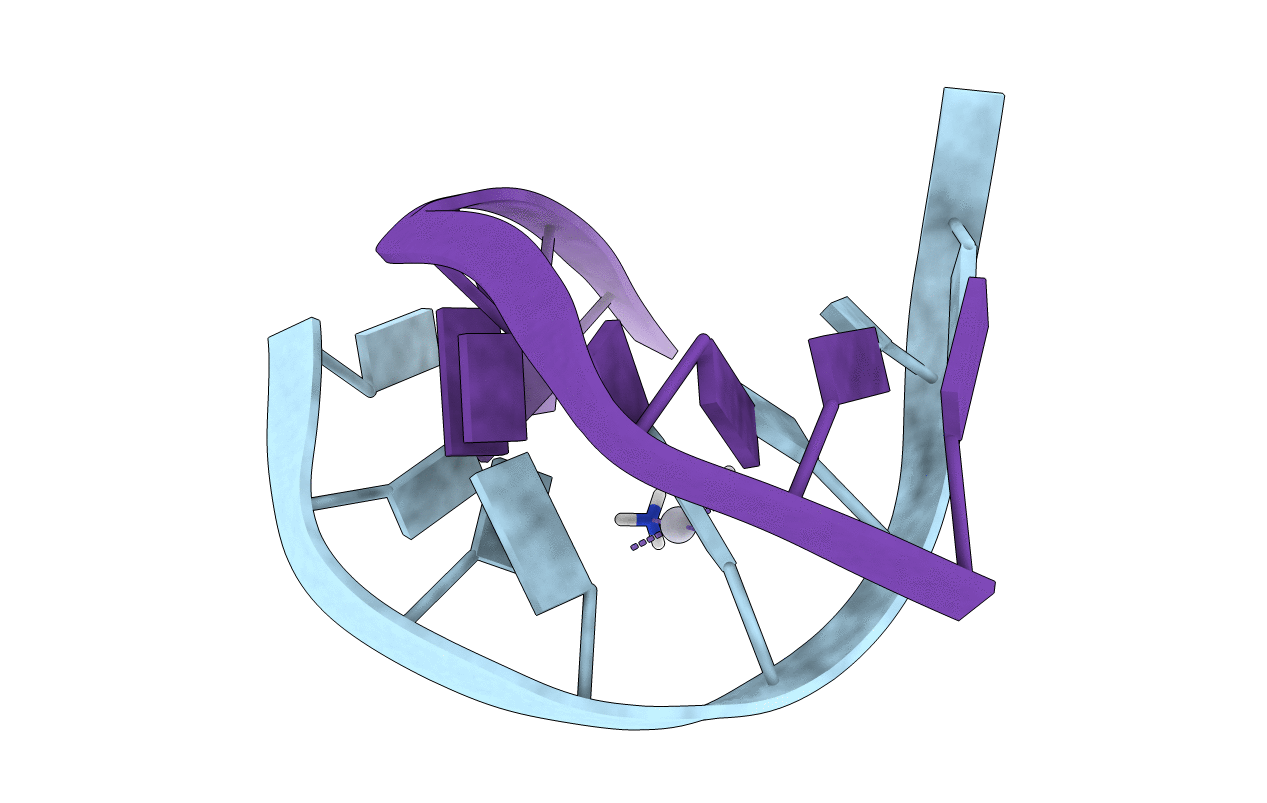
SOLUTION STRUCTURE OF INTRASTRAND CISPLATIN-CROSSLINKED DNA OCTAMER D(CCTG*G*TCC):D(GGACCAGG), NMR, MINIMIZED AVERAGE STRUCTURE
Method Details: