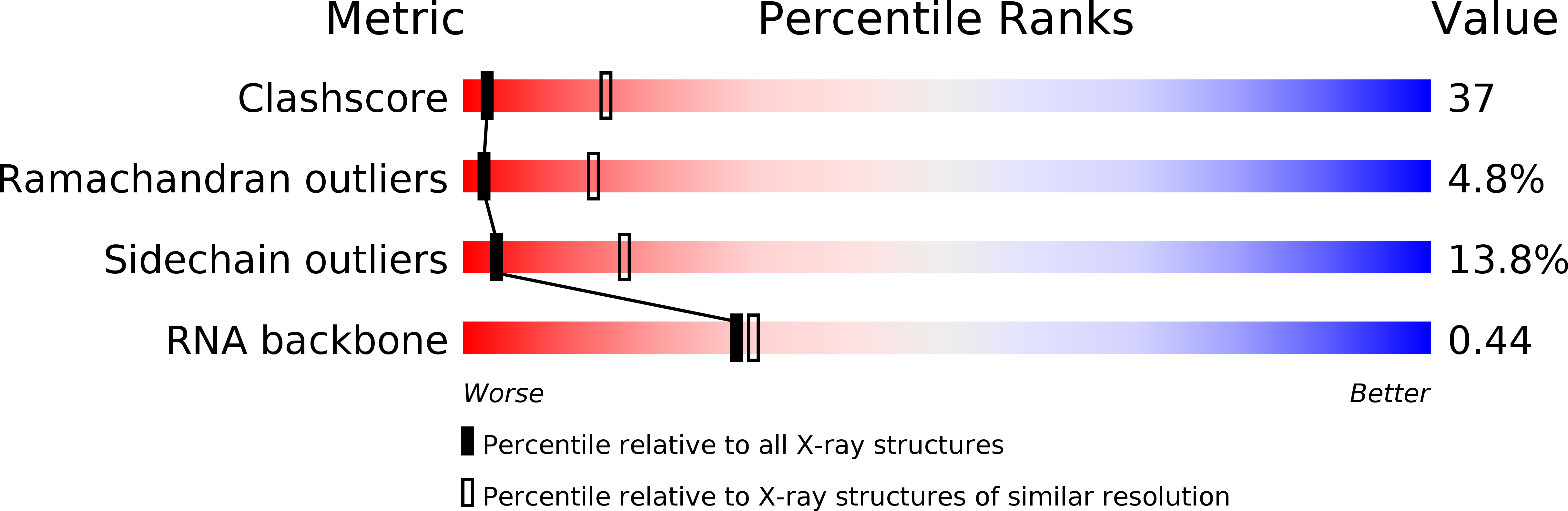
Deposition Date
1995-01-19
Release Date
1995-05-08
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1ASZ
Keywords:
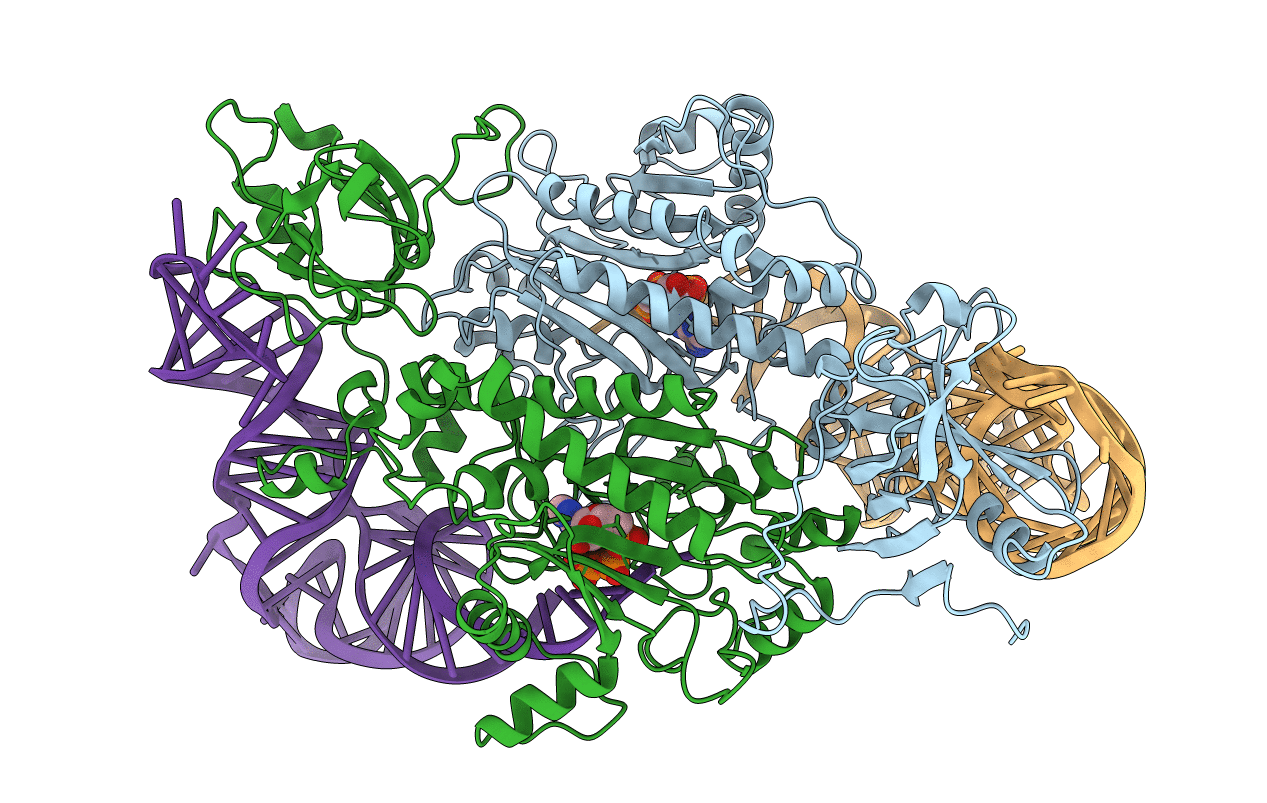
Title:
THE ACTIVE SITE OF YEAST ASPARTYL-TRNA SYNTHETASE: STRUCTURAL AND FUNCTIONAL ASPECTS OF THE AMINOACYLATION REACTION
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae (Taxon ID: )
Method Details:
Experimental Method:
Resolution:
3.00 Å
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 21 21 2