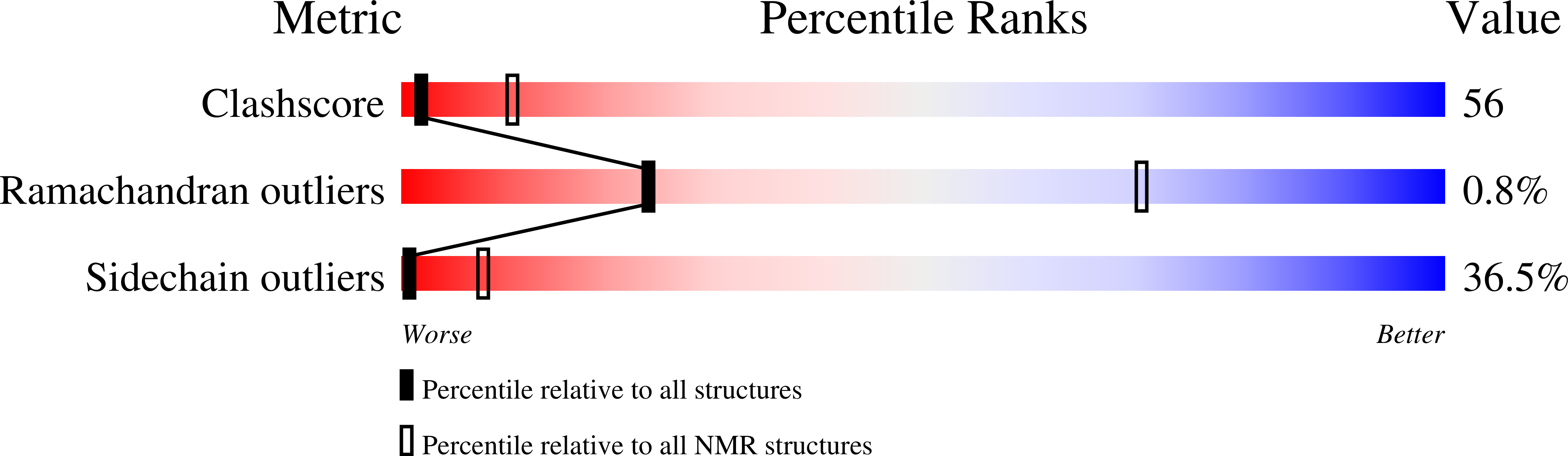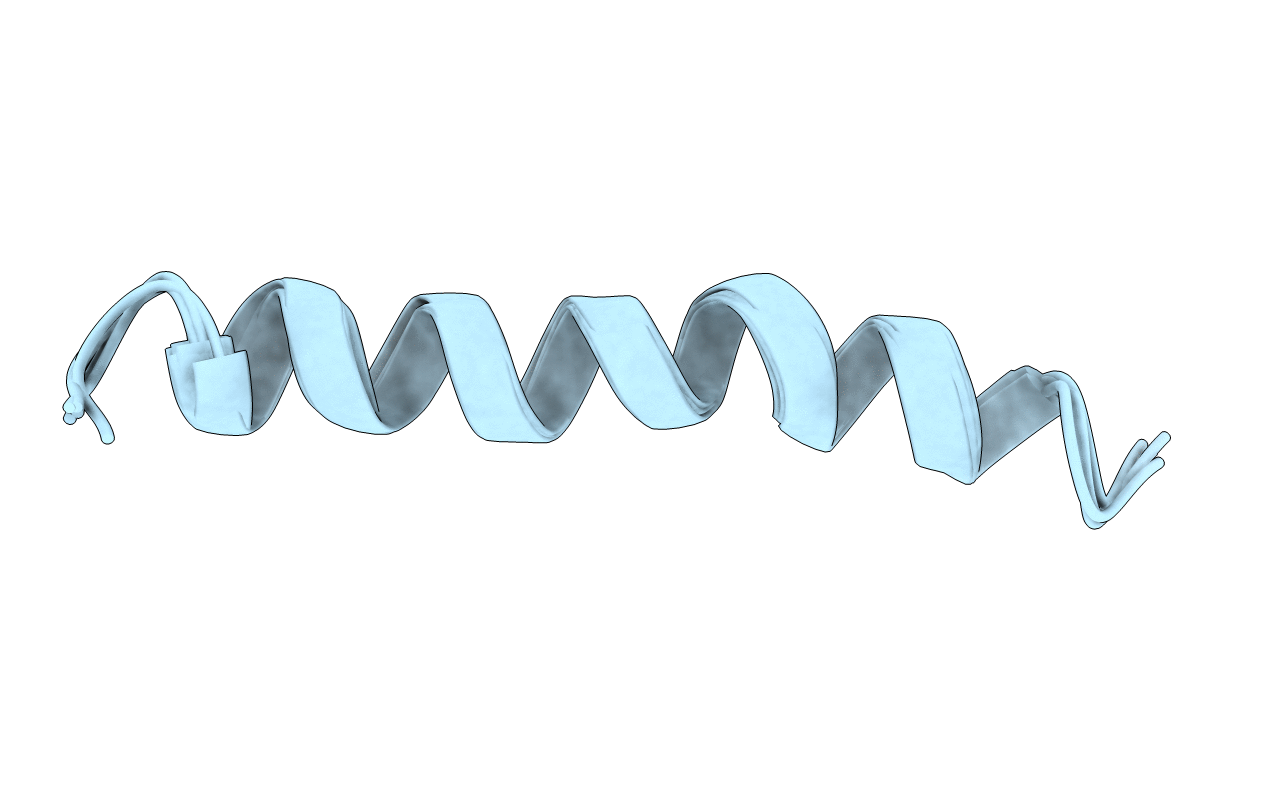
Deposition Date
1994-11-14
Release Date
1995-01-26
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1AMC
Keywords:
Title:
SOLUTION STRUCTURE OF RESIDUES 1-28 OF THE AMYLOID BETA-PEPTIDE
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Method Details:
Experimental Method:
Conformers Submitted:
5