Deposition Date
1998-02-17
Release Date
1998-05-27
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1A5N
Keywords:
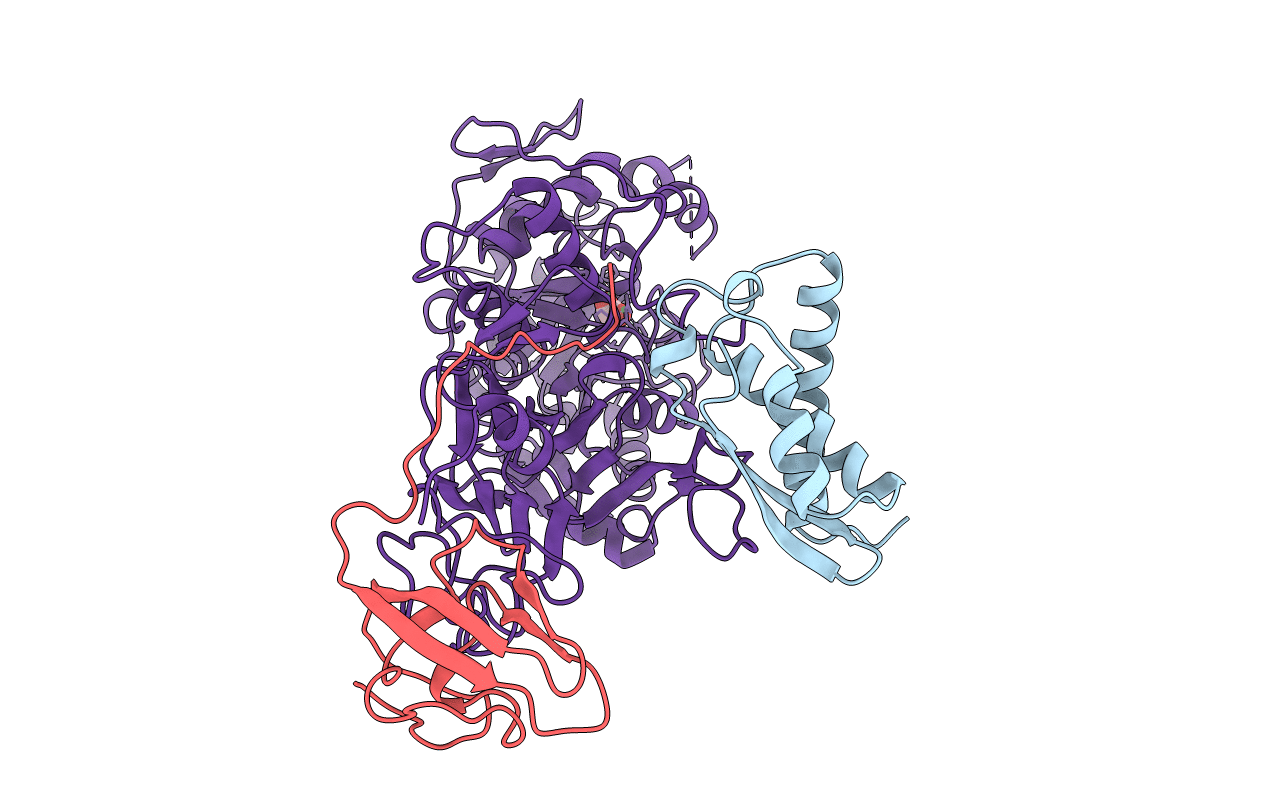
Title:
K217A VARIANT OF KLEBSIELLA AEROGENES UREASE, CHEMICALLY RESCUED BY FORMATE AND NICKEL
Biological Source:
Source Organism:
Klebsiella aerogenes (Taxon ID: 28451)
Host Organism:
Method Details:
Experimental Method:
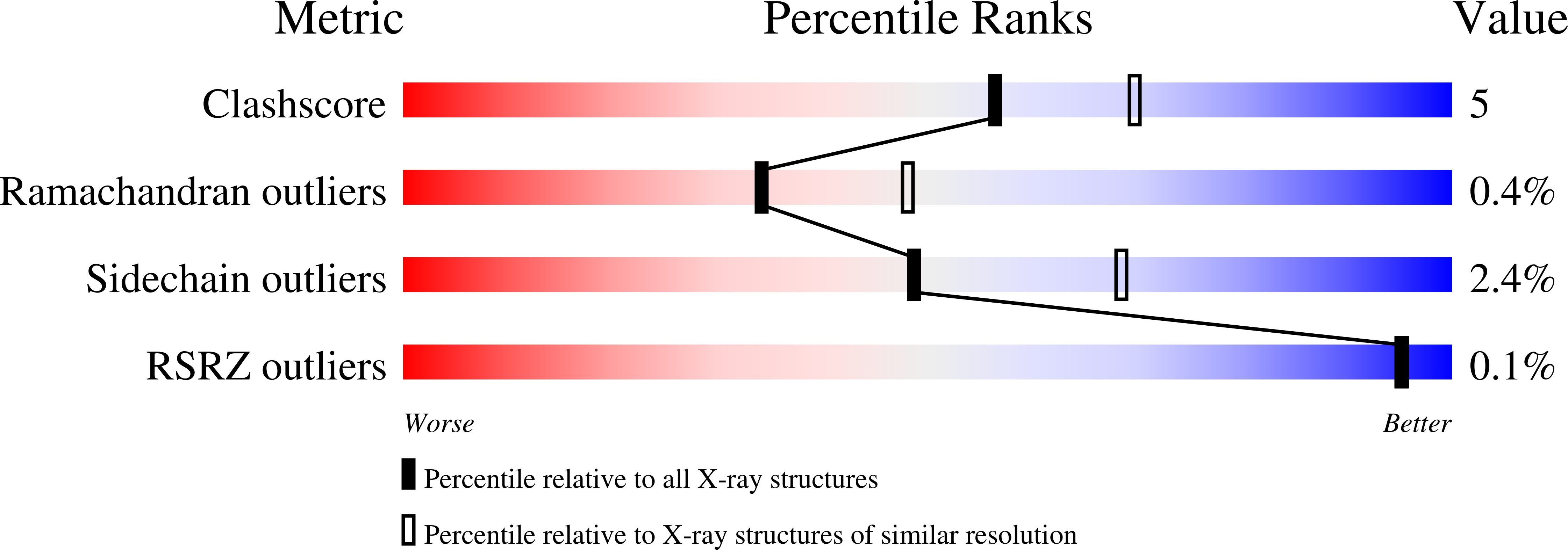
Resolution:
2.40 Å
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
I 21 3