Deposition Date
1994-08-22
Release Date
1994-11-01
Last Version Date
2024-05-22
Entry Detail
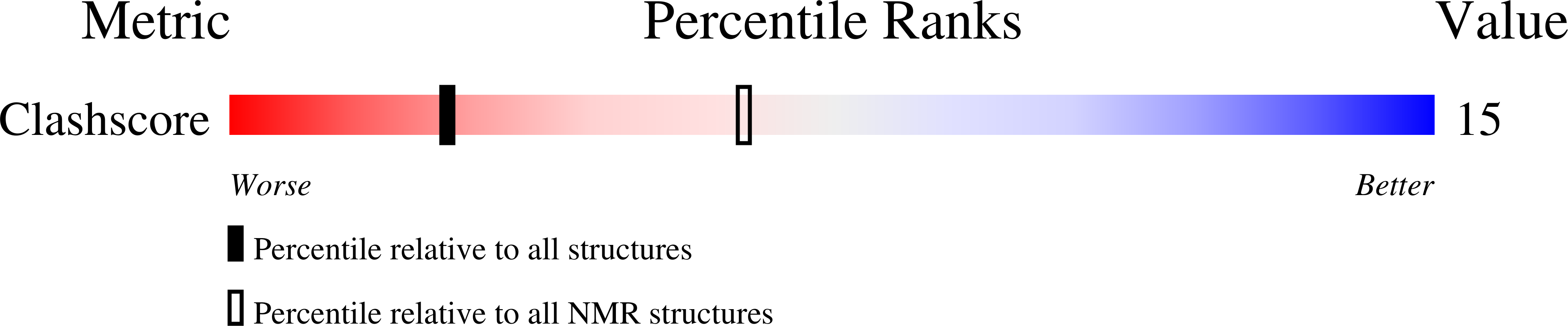
PDB ID:
186D
Keywords:
Title:
SOLUTION STRUCTURE OF THE TETRAHYMENA TELOMERIC REPEAT D(T2G4)4 G-TETRAPLEX
Method Details:
Experimental Method:
Conformers Submitted:
7