Deposition Date
1994-08-10
Release Date
1995-07-10
Last Version Date
2024-02-07
Entry Detail
PDB ID:
184D
Keywords:
Title:
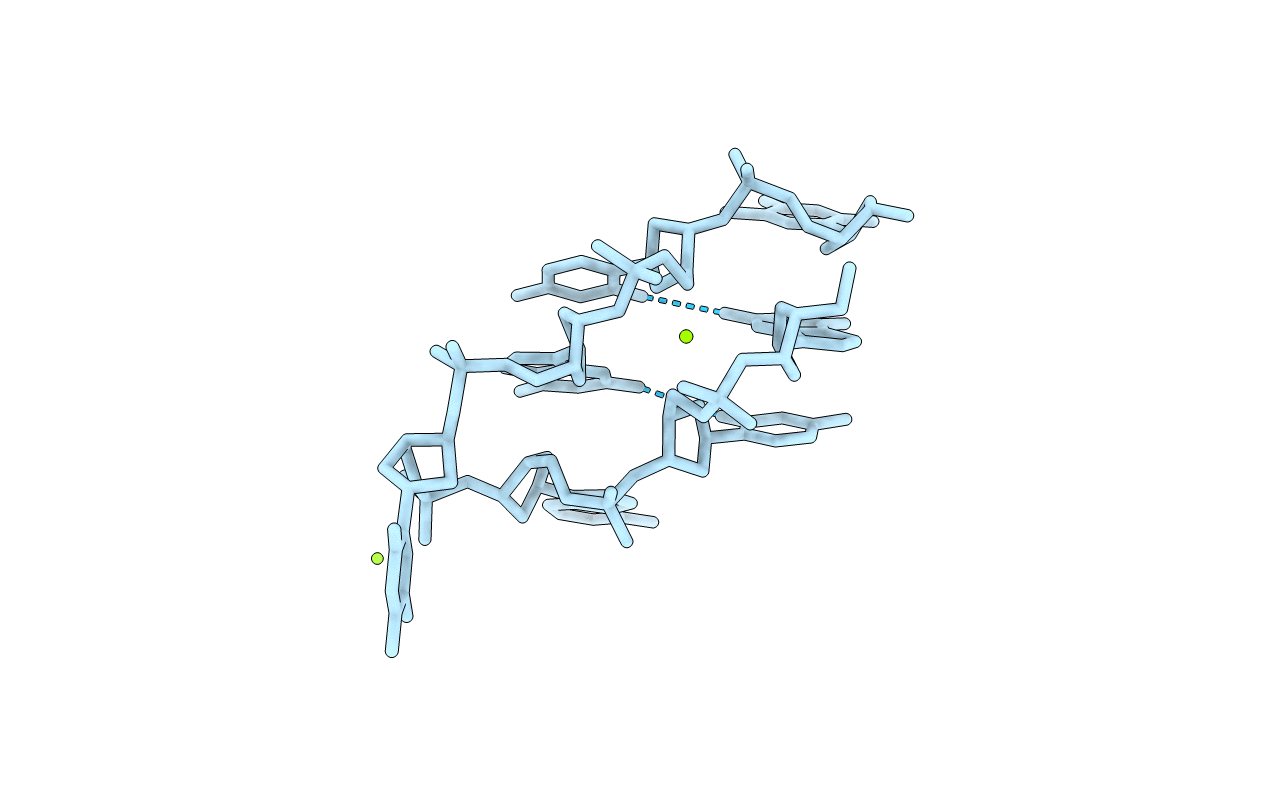
SELF-ASSOCIATION OF A DNA LOOP CREATES A QUADRUPLEX: CRYSTAL STRUCTURE OF D(GCATGCT) AT 1.8 ANGSTROMS RESOLUTION
Method Details:
Experimental Method:
Resolution:
1.80 Å
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
C 2 2 2