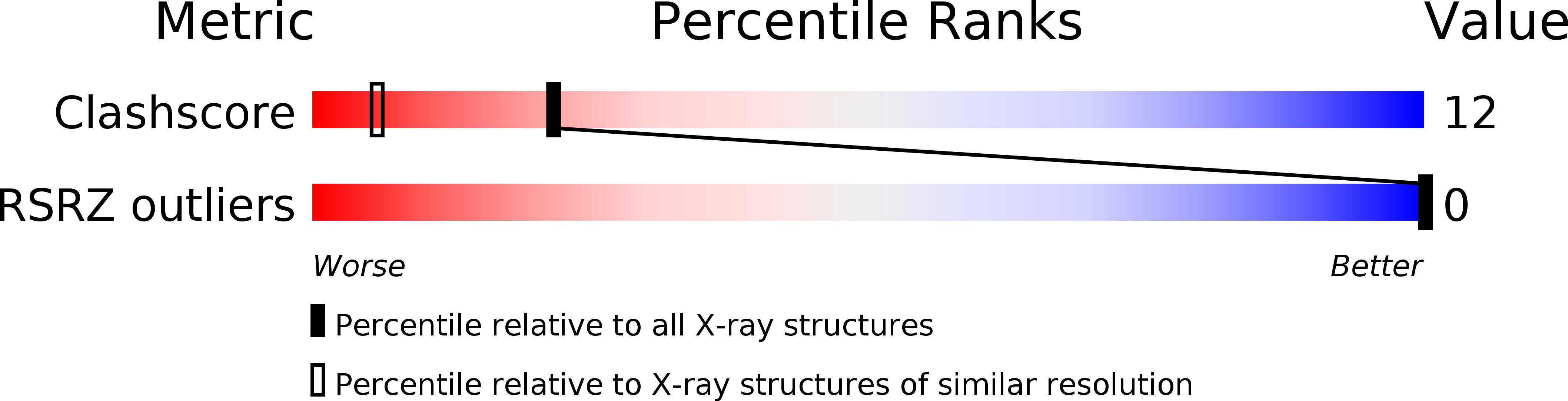
Deposition Date
1993-02-11
Release Date
1993-02-11
Last Version Date
2023-03-22
Entry Detail
PDB ID:
118D
Keywords:
Title:
CRYSTAL AND MOLECULAR STRUCTURE OF D(GTGCGCAC): INVESTIGATION OF THE EFFECTS OF BASE SEQUENCE ON THE CONFORMATION OF OCTAMER DUPLEXES
Method Details:
Experimental Method:
Resolution:
1.64 Å
R-Value Observed:
0.15
Space Group:
P 43 21 2