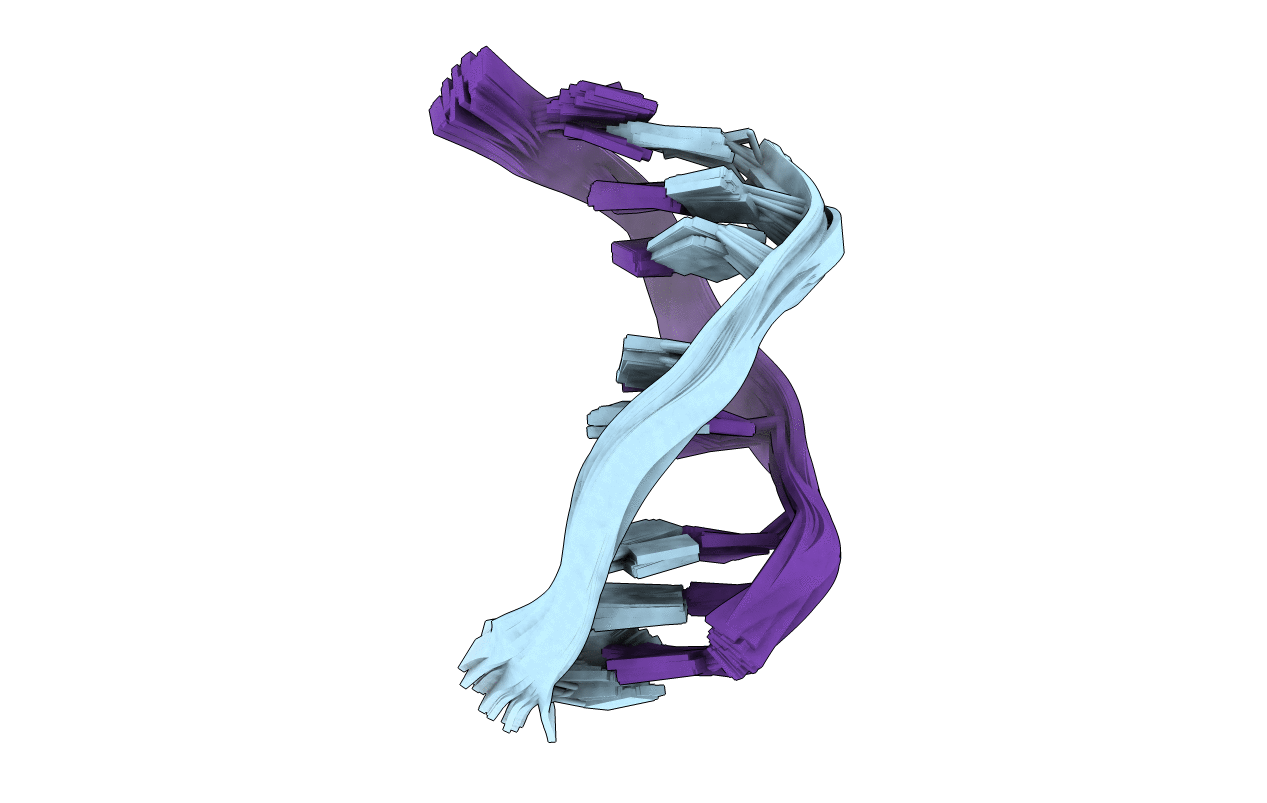
Deposition Date
1995-01-31
Release Date
1995-06-03
Last Version Date
2024-03-13
Entry Detail
PDB ID:
108D
Keywords:
Title:
THE SOLUTION STRUCTURE OF A DNA COMPLEX WITH THE FLUORESCENT BIS INTERCALATOR TOTO DETERMINED BY NMR SPECTROSCOPY
Method Details:
Experimental Method:
Conformers Submitted:
40


