Search Count: 147
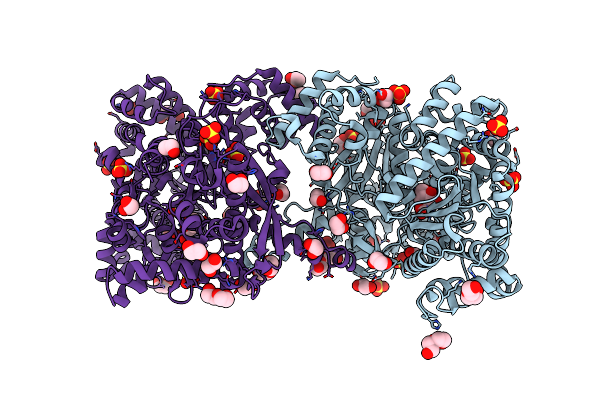 |
Maltodextrin Phosphorylase (Malp) In Complex With A Alpha-1,2-Cyclophellitol Analogue
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-05-15 Classification: TRANSFERASE Ligands: A1H3V, EDO, PEG, SO4 |
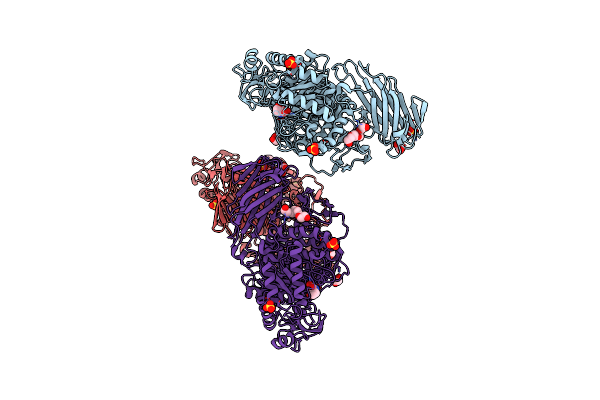 |
Crystal Structure Of Agd31B, Alpha-Transglucosylase, Complexed With A Non-Covalent 1,2- Cyclophellitol Aziridine
Organism: Cellvibrio japonicus ueda107
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-05-15 Classification: STRUCTURAL PROTEIN Ligands: GDQ, OXL, SO4, EDO, PG4 |
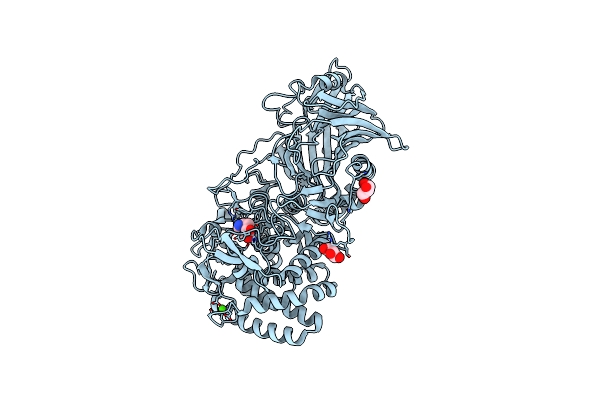 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With A Covalently Bound Cyclophellitol Aziridine
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-11-22 Classification: HYDROLASE Ligands: PO8, CA, GOL, CL |
 |
Organism: Cellvibrio japonicus ueda107
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-04 Classification: HYDROLASE |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-08-09 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-08-09 Classification: IMMUNE SYSTEM Ligands: CA, L6N |
 |
Crystal Structure Of Agd31B, Alpha-Transglucosylase In Glycoside Hydrolase Family 31, In Complex With Noncovalent Cyclophellitol Sulfamidate Probe Kk131
Organism: Cellvibrio japonicus (strain ueda107)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2022-07-27 Classification: HYDROLASE Ligands: EDO, OXL, 5OV, SO4 |
 |
Crystal Structure Of Agd31B, Alpha-Transglucosylase In Glycoside Hydrolase Family 31, In Complex With Covalent Cyclophellitol Sulfamidate Probe Kk130
Organism: Cellvibrio japonicus ueda107
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-07-27 Classification: HYDROLASE Ligands: EDO, PG4, OXL, 56I, SO4 |
 |
Crystal Structure Of Human Lysosomal Acid-Alpha-Glucosidase, Gaa, In Complex With Cyclosulfamidate 4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: SO4, CL, GOL, EDO, YTW, PGE |
 |
Crystal Structure Of Human Lysosomal Acid-Alpha-Glucosidase, Gaa, In Complex With Cyclosulfamidate 6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: NAG, 56I, SO4, CL, GOL, EDO, PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2021-11-17 Classification: LIPID BINDING PROTEIN, IMMUNE SYSTEM Ligands: NAG, ZPD, D12, ZP7, NHE, MLA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-11-17 Classification: LIPID BINDING PROTEIN, IMMUNE SYSTEM Ligands: NAG, NHE, ZQ4, D12, ZP7, MLA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2021-11-17 Classification: LIPID BINDING PROTEIN, IMMUNE SYSTEM Ligands: NAG, ZQ7, D12, ZP7, MLA |
 |
Human Adp-Ribosylserine Hydrolase Arh3 Mutant E41A In Complex With Adp-Ribose Dimer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-06-16 Classification: HYDROLASE Ligands: MG, AR6, CL, EDO |
 |
Human Adp-Ribosylserine Hydrolase Arh3 Mutant E41A In Complex With H2B-S7-Mar Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2021-06-16 Classification: HYDROLASE Ligands: MG, EDO, ACT, AR6 |
 |
Adp-Ribosylserine Hydrolase Arh3 Of Latimeria Chalumnae In Complex With Alpha-1''-O-Methyl-Adp-Ribose (Meadpr)
Organism: Latimeria chalumnae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-16 Classification: HYDROLASE Ligands: MG, RVK |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2021-06-16 Classification: HYDROLASE Ligands: MG, 8NA, EDO, ACT |
 |
Structure Of The D125N Mutant Of The Catalytic Domain Of The Bacillus Circulans Alpha-1,6 Mannanase In Complex With An Alpha-1,6-Alpha-Manno-Cyclophellitol Carbasugar-Stabilised Trisaccharide Inhibitor
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2021-04-28 Classification: HYDROLASE Ligands: EDO, QE8, MAN |
 |
Structure Of The D125N Mutant Of The Catalytic Domain Of The Bacillus Circulans Alpha-1,6 Mannanase In Complex With An Alpha-1,6-Alpha-Manno-Cyclophellitol Trisaccharide Inhibitor
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-04-28 Classification: HYDROLASE Ligands: PBW, EDO |
 |
Structure Of The Catalytic Domain Of The Bacillus Circulans Alpha-1,6 Mannanase In Complex With An Alpha-1,6- Alpha-Manno-Cyclophellitol Carbasugar-Stabilised Trisaccharide Inhibitor
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-04-28 Classification: HYDROLASE Ligands: EDO, PBW, M96, MAN |

