Search Count: 79
 |
Inhibiting The Virulence Of Gut Bacteria Through Blocking The Activation Of A Two-Component Lanthipeptide Toxin
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE |
 |
Solution Nmr Structure Of The Ripp Proteusin Dpre2 From Desulfotomaculum Sp.
Organism: Desulfotomaculum sp.
Method: SOLUTION NMR Release Date: 2024-12-04 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of A Holoenzyme Fe-Free Tglhi For Pseudomonas Syringae Peptidyl (S) 2-Mercaptoglycine Biosynthesis
Organism: Pseudomonas syringae pv. maculicola str. es4326
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-08-23 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of A Holoenzyme Tglhi With Two Fe Irons For Pseudomonas Syringae Peptidyl (S) 2-Mercaptoglycine Biosynthesis
Organism: Pseudomonas syringae pv. maculicola str. es4326
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2023-08-23 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Crystal Structure Of A Holoenzyme Tglhi With Three Fe Ions For Pseudomonas Syringae Peptidyl (S) 2-Mercaptoglycine Biosynthesis
Organism: Pseudomonas syringae pv. maculicola str. es4326
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2023-08-23 Classification: METAL BINDING PROTEIN Ligands: FE |
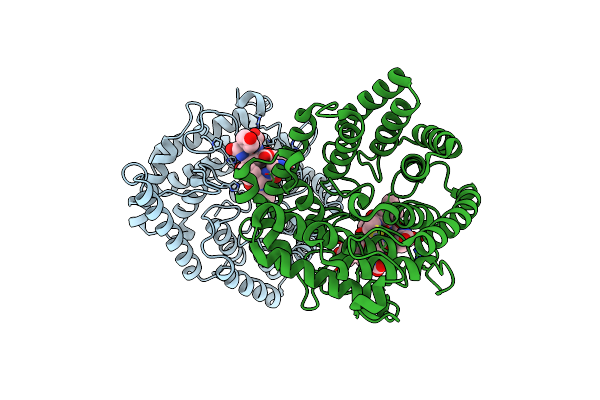 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-01-25 Classification: SIGNALING PROTEIN Ligands: ZN, GSH |
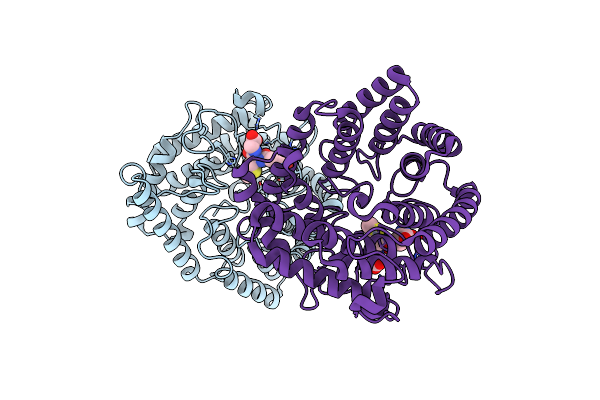 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-01-25 Classification: SIGNALING PROTEIN Ligands: ZN, GSM |
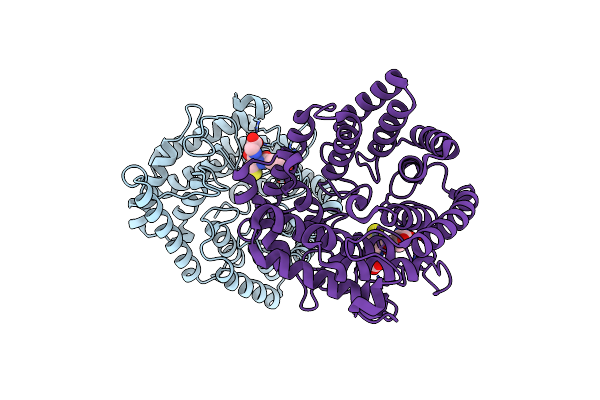 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2023-01-25 Classification: SIGNALING PROTEIN Ligands: ZN, GSH |
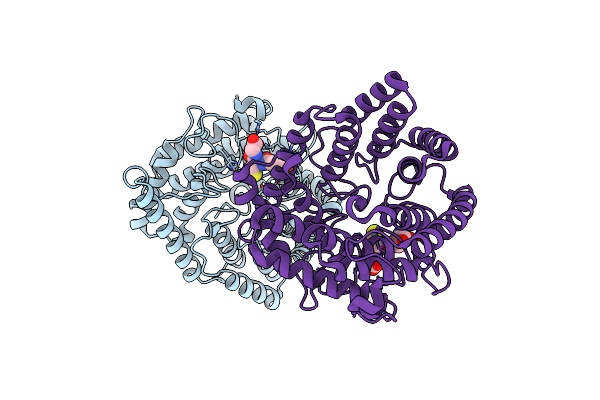 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2023-01-25 Classification: SIGNALING PROTEIN Ligands: ZN, GSH |
 |
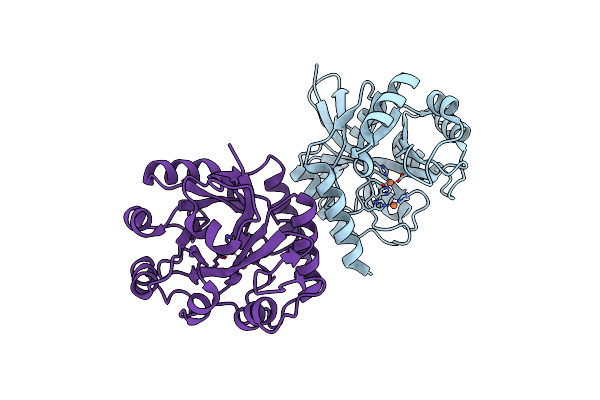
Organism: Streptomyces morookaense
Method: SOLUTION NMR Release Date: 2022-10-12 Classification: BIOSYNTHETIC PROTEIN |
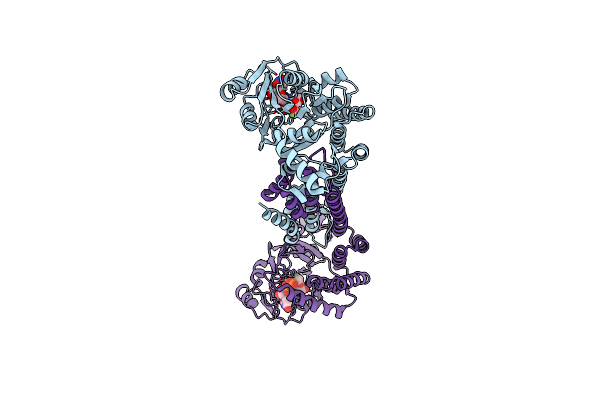 |
Organism: Bacillus thuringiensis serovar andalousiensis bgsc 4aw1
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2022-04-13 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, U2F |
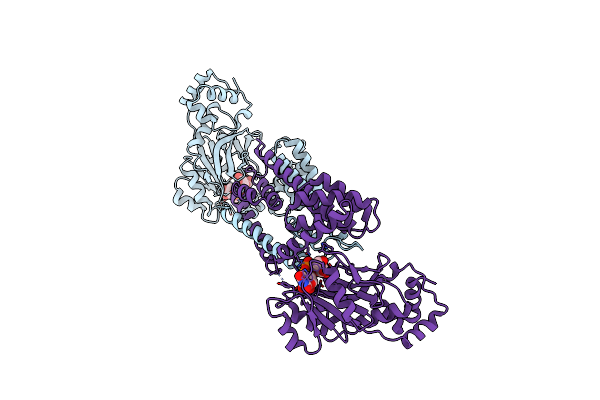 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-04-13 Classification: TRANSFERASE Ligands: UPG |
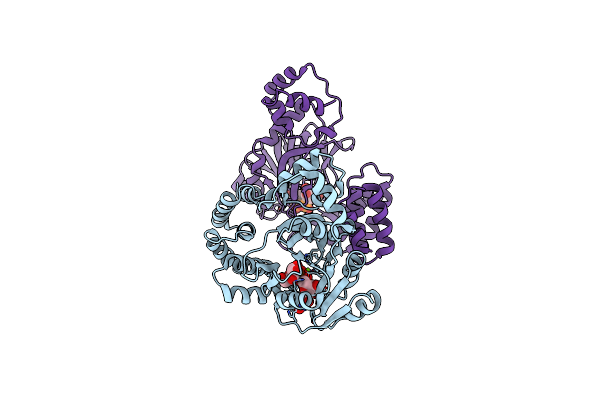 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-04-13 Classification: BIOSYNTHETIC PROTEIN Ligands: UPG, MG |
 |
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2021-09-15 Classification: BIOSYNTHETIC PROTEIN Ligands: FE2 |
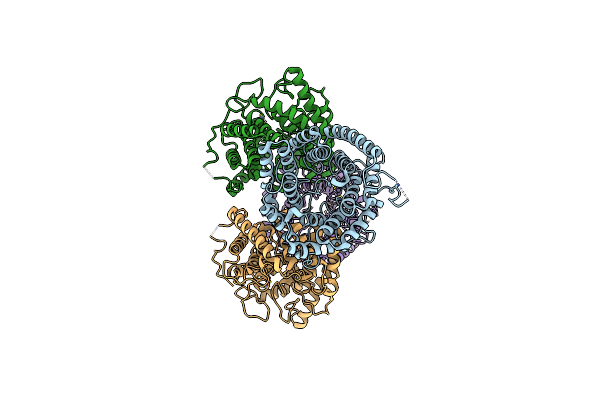 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-05-05 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN |
 |
Solution Nmr Structure Of Prochlorosin 2.11 (Pcn2.11) Produced By Prochlorococcus Mit 9313
Organism: Prochlorococcus marinus str. mit 9313
Method: SOLUTION NMR Release Date: 2020-09-09 Classification: UNKNOWN FUNCTION |
 |
Solution Nmr Structure Of Prochlorosin 2.10 Produced By Prochlorococcus Mit 9313
Organism: Prochlorococcus marinus str. mit 9313
Method: SOLUTION NMR Release Date: 2020-09-09 Classification: UNKNOWN FUNCTION |
 |
Solution Nmr Structure Of Enterococcal Cytolysin S (Cylls") Produced By Enterococcus Faecalis
|
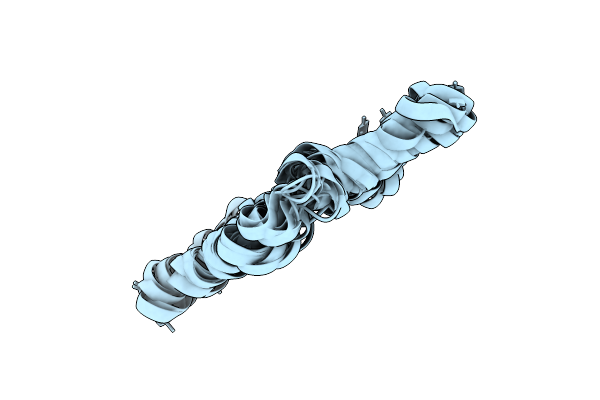 |
Solution Nmr Structure Of Enterococcal Cytolysin L (Cylll") Produced By Enterococcus Faecalis
|
 |
Organism: Prochlorococcus marinus str. mit 9313
Method: SOLUTION NMR Release Date: 2020-07-08 Classification: UNKNOWN FUNCTION |

