Search Count: 7
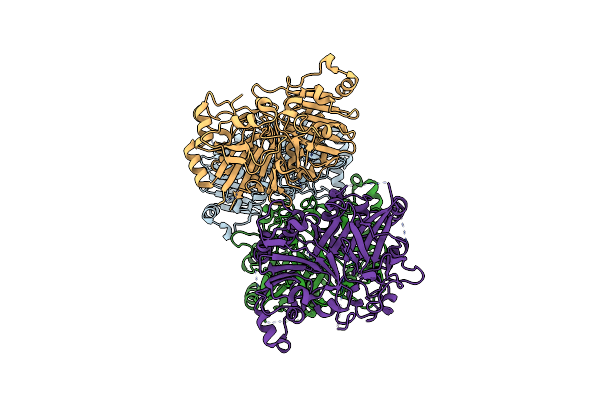 |
Crystal Structure Analysis Of The Yeast Tyrosyl-Dna Phosphodiesterase H182A Mutant
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-12-28 Classification: HYDROLASE |
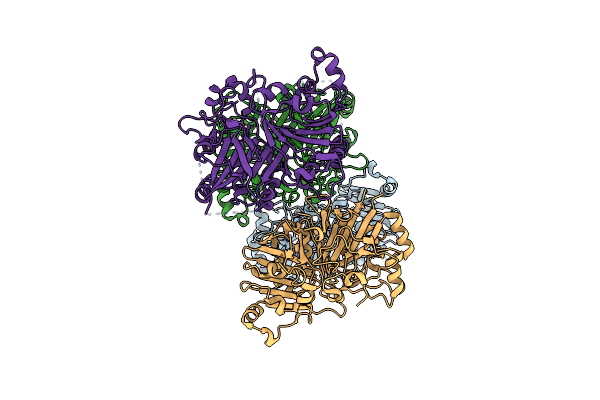 |
Crystal Structure Analysis Of The Yeast Tyrosyl-Dna Phosphodiesterase H432N Mutant
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-12-28 Classification: HYDROLASE |
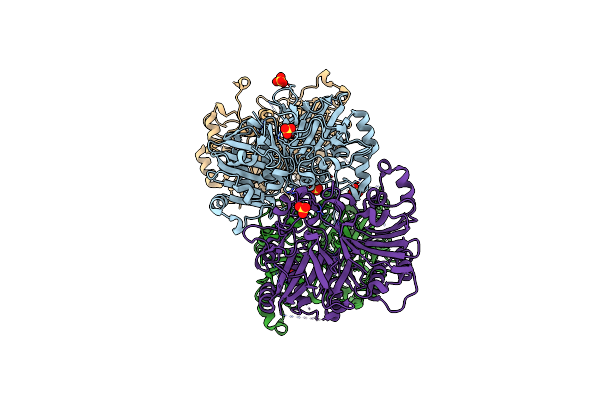 |
Crystal Structure Analysis Of The Yeast Tyrosyl-Dna Phosphodiesterase H432N_Glu Mutant
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-12-28 Classification: HYDROLASE Ligands: SO4 |
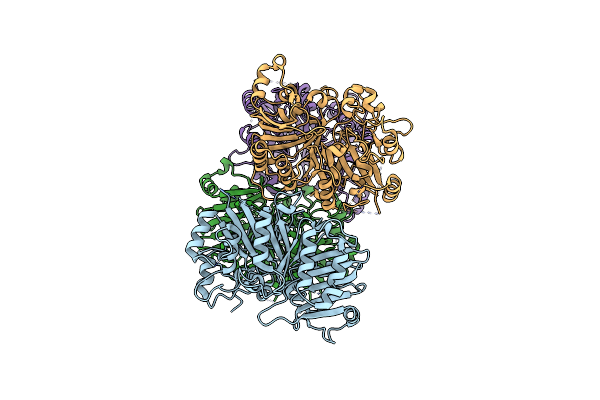 |
Crystal Structure Analysis Of The Yeast Tyrosyl-Dna Phosphodiesterase 1 H432R Mutant (Scan1 Mutant)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-12-28 Classification: HYDROLASE |
 |
Structure Of A Sumo-Binding-Motif Mimic Bound To Smt3P-Ubc9P: Conservation Of A Noncovalent Ubiquitin-Like Protein-E2 Complex As A Platform For Selective Interactions Within A Sumo Pathway
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-05-29 Classification: LIGASE/PROTEIN BINDING |
 |
Distinct Functional Domains Of Ubc9 Dictate Cell Survival And Resistance To Genotoxic Stress
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2006-07-04 Classification: LIGASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2004-09-07 Classification: REPLICATION,TRANSCRIPTION,HYDROLASE |

