Search Count: 31
 |
Crystal Structure Of Micobacterium Tuberculosis Udp-Galactopyranose Mutase In Complex With Substrate Udp-Galp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-01-21 Classification: ISOMERASE Ligands: FAD, GDU, UDP |
 |
Crystal Structure Of Micobacterium Tuberculosis Udp-Galactopyranose Mutase In Complex With Substrate Udp-Galp (Reduced)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-01-21 Classification: ISOMERASE Ligands: FAD, GDU |
 |
Crystal Structure Of Micobacterium Tuberculosis Udp-Galactopyranose Mutase In Complex With Udp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-01-21 Classification: ISOMERASE Ligands: FAD, UDP |
 |
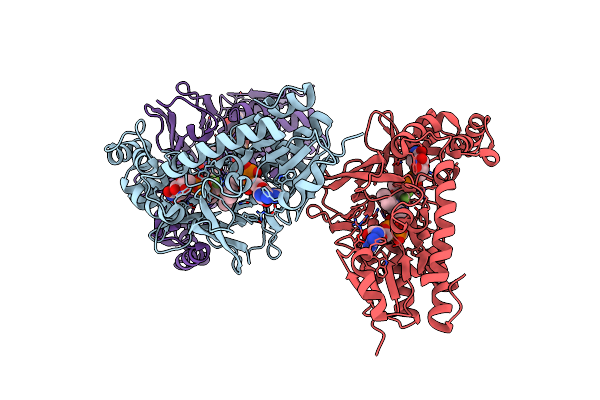
Crystal Structure Of Micobacterium Tuberculosis Udp-Galactopyranose Mutase In Complex With Tetrafluorinated Substrate Analog Udp-F4-Galf
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2015-01-21 Classification: ISOMERASE Ligands: FAD, 3UA |
 |
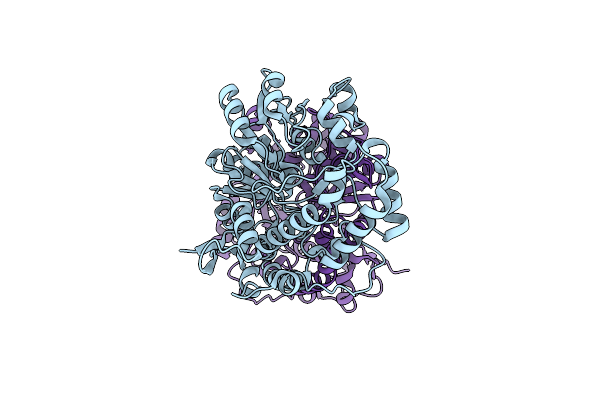
Crystal Structure Of Micobacterium Tuberculosis Udp-Galactopyranose Mutase In Complex With Tetrafluorinated Substrate Analog Udp-F4-Galp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-01-21 Classification: ISOMERASE Ligands: FAD, 3UC |
 |
Crystal Structure Of Ntda From Bacillus Subtilis In Complex With The Internal Aldimine
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2013-10-16 Classification: TRANSFERASE |
 |
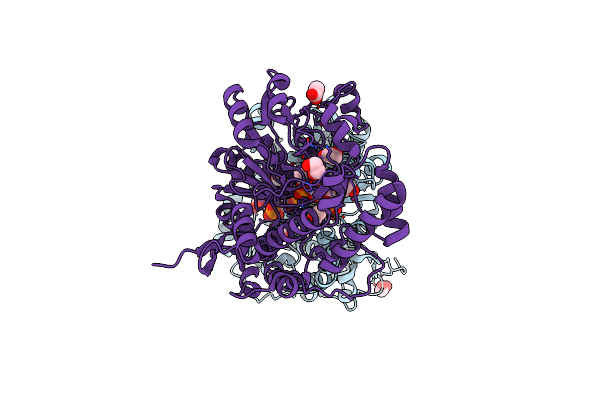
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2013-10-16 Classification: TRANSFERASE Ligands: PMP, ACT, EDO, CL |
 |
Crystal Structure Of Ntda From Bacillus Subtilis In Complex With The Plp External Aldimine Adduct With Kanosamine-6-Phosphate
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2013-10-16 Classification: TRANSFERASE Ligands: O1G, EDO, ACT |
 |
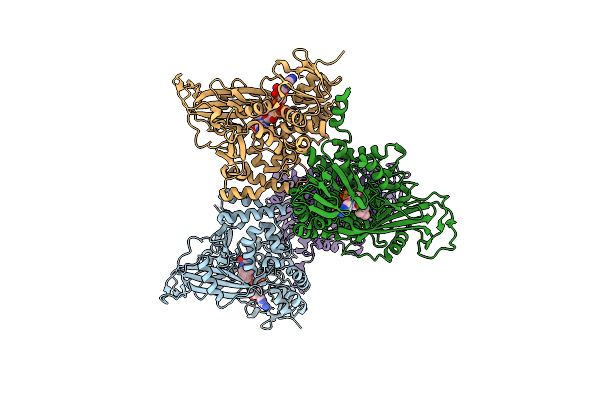
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2012-08-22 Classification: SIGNALING PROTEIN, HORMONE Ligands: PO4, CL |
 |
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2012-02-22 Classification: ISOMERASE Ligands: FAD |
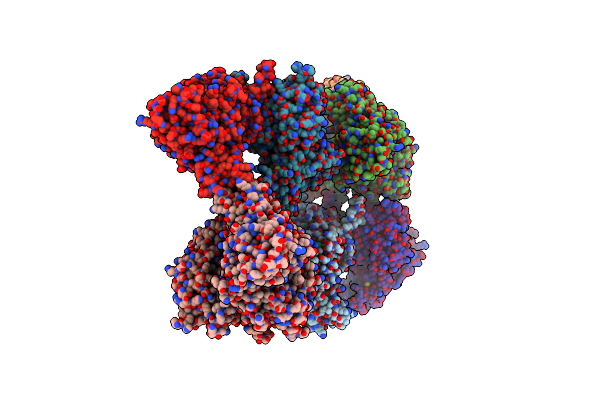 |
Crystal Structure Of Udp-Galactopyranose Mutase From Aspergillus Fumigatus In Complex With Udpgalp (Reduced)
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-02-22 Classification: ISOMERASE Ligands: GDU, FDA, CL |
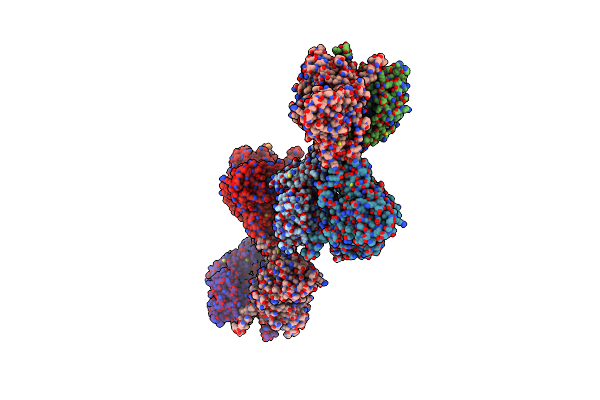 |
Crystal Structure Of Udp-Galactopyranose Mutase From Aspergillus Fumigatus In Complex With Udpgalp (Non-Reduced)
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-02-22 Classification: ISOMERASE Ligands: FAD, GDU, CL, FDA |
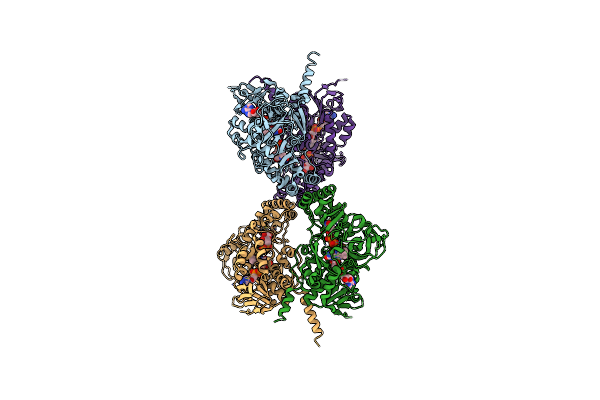 |
Crystal Structure Of R182K-Udp-Galactopuranose Mutase From Aspergillus Fumigatus In Complex With Udpgalp
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2012-02-22 Classification: ISOMERASE Ligands: FDA, UDP, FAD |
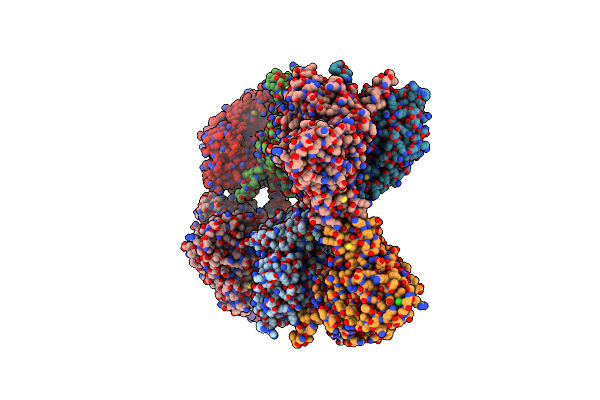 |
Crystal Structure Of Udp-Galactopyranose Mutase From Aspergillus Fumigatus In Complex With Udp
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2012-02-22 Classification: ISOMERASE Ligands: UDP, FAD, CL |
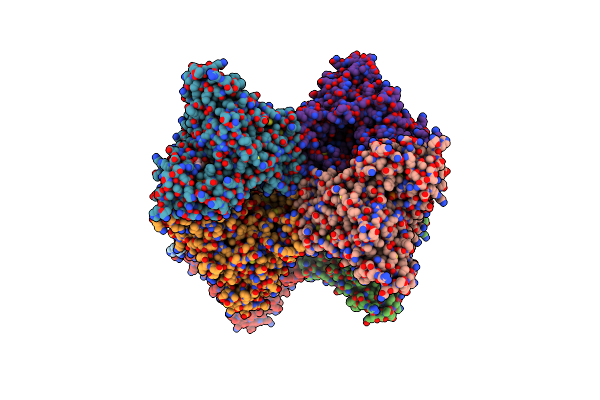 |
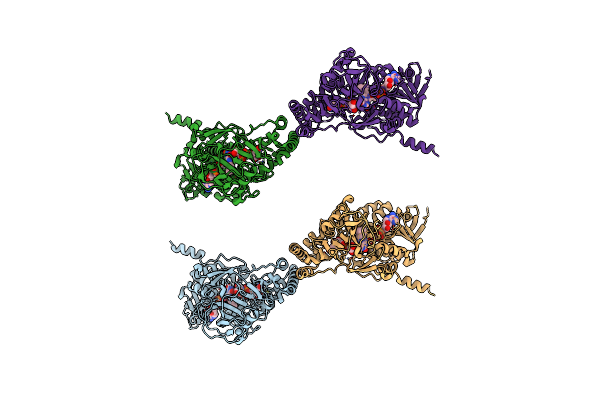
Crystal Structure Of R327A Udp-Galactopyranose Mutase From Aspergillus Fumigatus In Complex With Udpgalp
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2012-02-22 Classification: ISOMERASE Ligands: FAD, GDU |
 |
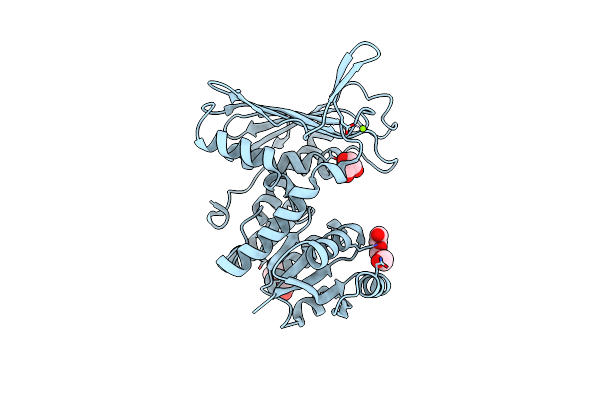
Crystal Structure Of R327K Udp-Galactopyranose Mutase From Aspergillus Fumigatus In Complex With Udpgalp
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2012-02-22 Classification: ISOMERASE Ligands: FAD, GDU |
 |
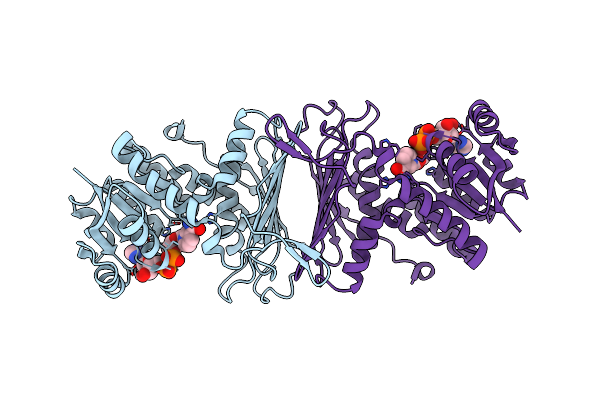
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2010-09-29 Classification: OXIDOREDUCTASE Ligands: PGE, GOL, CL, MG |
 |
Crystal Structure Of Myo-Inositol Dehydrogenase From Bacillus Subtilis With Bound Cofactor
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-09-15 Classification: OXIDOREDUCTASE Ligands: NAD, NAI |
 |
Crystal Structure Of Myo-Inositol Dehydrogenase From Bacillus Subtilis With Bound Cofactor Nadh And Inositol
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-09-15 Classification: OXIDOREDUCTASE Ligands: NAI, INS |
 |
Crystal Structure Of Myo-Inositol Dehydrogenase From Bacillus Subtilis With Bound Cofactor And Product Inosose
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-09-15 Classification: OXIDOREDUCTASE Ligands: NAD, ISE, NAI |

