Search Count: 127
 |
Structure Of The Bacteriophage K Gp155 C-Terminal Domain, Seleno-Methionine Modified Version
Organism: Staphylococcus phage k
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: G3P, CA, PO4 |
 |
Cryo-Em Structure Of Staphylococcus Aureus Bacteriophage Phi812 Baseplate In The Pre-Contraction State - Complete
Organism: Staphylococcus phage 812
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: VIRUS |
 |
Cryo-Em Structure Of Staphylococcus Aureus Bacteriophage Phi812 Baseplate In The Pre-Contraction State - Core, Wedge Module, And Proximal Tail Proteins
Organism: Staphylococcus phage 812
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: VIRUS |
 |
Cryo-Em Structure Of Staphylococcus Aureus Bacteriophage Phi812 Baseplate In The Pre-Contraction State - Core, And Wedge Module Proteins
Organism: Staphylococcus phage 812
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: VIRUS |
 |
Cryo-Em Structure Of Staphylococcus Aureus Bacteriophage Phi812 Baseplate In The Post-Contraction State - Complete
Organism: Staphylococcus phage 812
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: VIRUS |
 |
Cryo-Em Structure Of Staphylococcus Aureus Bacteriophage Phi812 Baseplate In The Post-Contraction State - Sheath Initiator, Wedge Module, Inner Tripod, Arm Segment, And Proximal Tail Sheath Proteins
Organism: Staphylococcus phage 812
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: VIRUS |
 |
Cryo-Em Structure Of Staphylococcus Aureus Bacteriophage Phi812 Baseplate In The Post-Contraction State - Sheath Initiator, Wedge Module, And Inner Tripod Proteins
Organism: Staphylococcus phage 812
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: VIRUS |
 |
Cryo-Em Structure Of Staphylococcus Aureus Bacteriophage Phi812 Central Spike Protein - Knob And Petal Domains
Organism: Staphylococcus phage 812
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: VIRAL PROTEIN |
 |
Organism: Staphylococcus phage k
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-04-09 Classification: VIRAL PROTEIN Ligands: MES, GOL |
 |
Crystal Structure Of Type I Dehydroquinase From Staphylococcus Aureus Inhibited By A Hydroxylamine Derivative
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-02-15 Classification: LYASE Ligands: PVI, SO4, CL |
 |
Crystal Structure Of Type I Dehydroquinase From Salmonella Typhi Inhibited By A Hydroxylamine Derivative
Organism: Salmonella enterica subsp. enterica serovar typhi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-02-15 Classification: LYASE Ligands: PVI, NA |
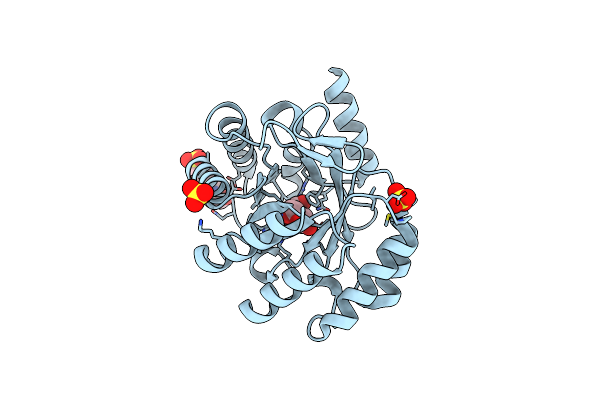 |
Crystal Structure Of Type I Dehydroquinase From Salmonella Typhi Inhibited By An Epoxide Derivative
Organism: Salmonella enterica subsp. enterica serovar typhi
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-02-15 Classification: LYASE Ligands: OVU, EPE, SO4 |
 |
Recognition Of Staphylococcus Aureus Wall Teichoic Acid Analogue Sa533 (Compound 1) By Fab4461
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-08-31 Classification: CARBOHYDRATE Ligands: 3CX, PEG, G6N |
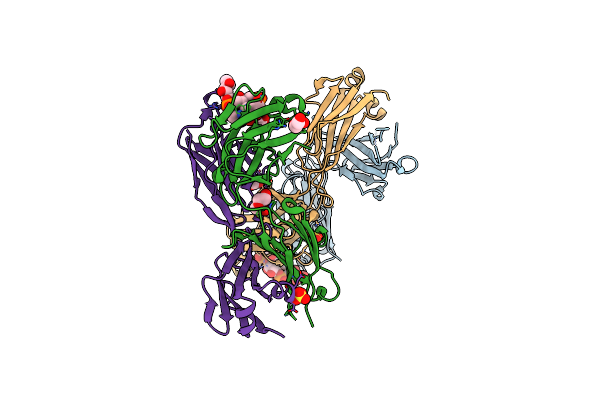 |
Recognition Of Staphylococcus Aureus Wall Teichoic Acid Analogue Sa475 (Compound 2) By Fab4497
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-08-31 Classification: CARBOHYDRATE Ligands: G5F, GOL, SO4, PEG |
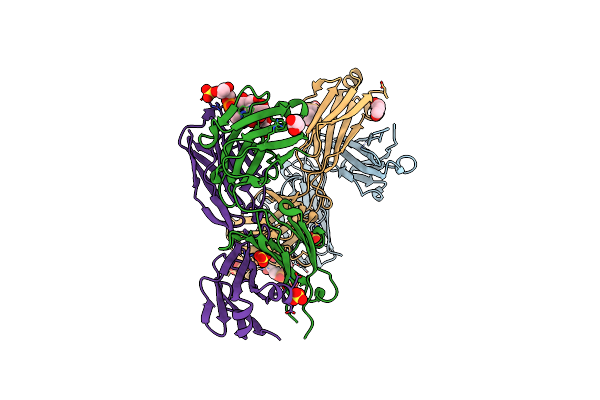 |
Recognition Of Staphylococcus Aureus Wall Teichoic Acid Analogue Tb87 (Compound 3) By Fab4497
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-08-31 Classification: CARBOHYDRATE Ligands: G7C, PEG, CL, SO4, GOL |
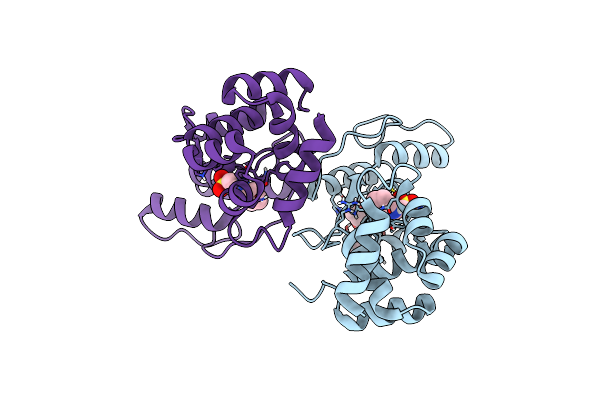 |
Structure Of The Pseudomonas Aeruginosa Bacteriophage Jg004 Endolysin Pae87, Apo Form.
Organism: Pseudomonas phage jg004
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-02-23 Classification: HYDROLASE Ligands: NHE, PG4 |
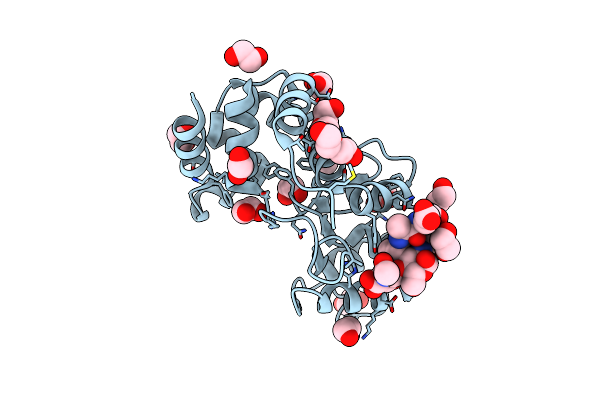 |
Structure Of The Pseudomonas Aeruginosa Bacteriophage Jg004 Endolysin Pae87 Bound To A Peptidoglycan Fragment.
Organism: Pseudomonas phage jg004, Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2022-02-23 Classification: HYDROLASE Ligands: PEG, EDO |
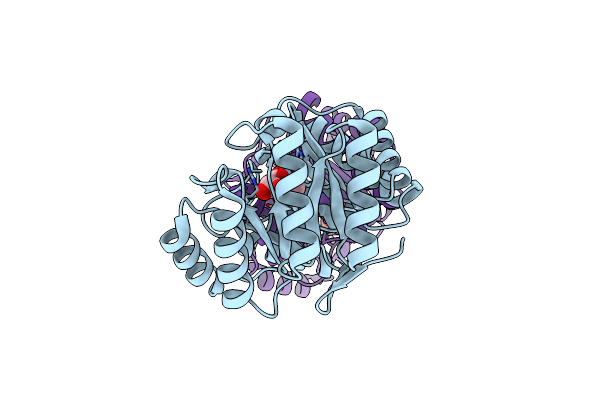 |
Crystal Structure Of Dhq1 From Salmonella Typhi Covalently Modified By Compound 7
Organism: Salmonella typhi
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2020-04-15 Classification: LYASE Ligands: L9Z |
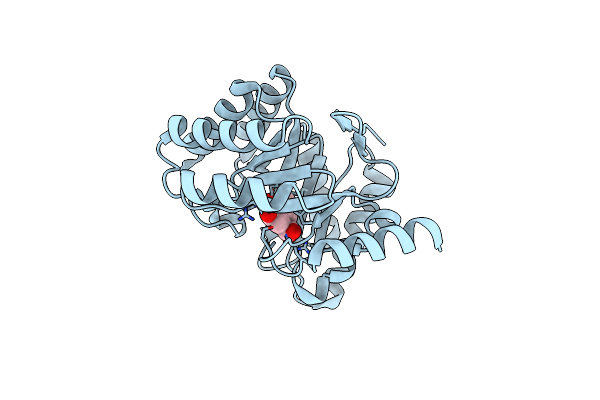 |
Crystal Structure Of Dhq1 From Salmonella Typhi Covalently Modified By Compound 9
Organism: Salmonella typhi
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2020-04-15 Classification: LYASE Ligands: FQZ, FSQ |
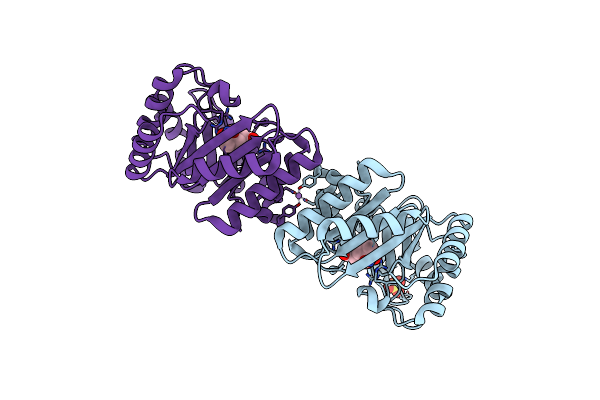 |
Crystal Structure Of Dhq1 From Staphylococcus Aureus Covalently Modified By Ligand 7
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2020-04-15 Classification: LYASE Ligands: L9Z, SO4, LI |

