Search Count: 44,349
 |
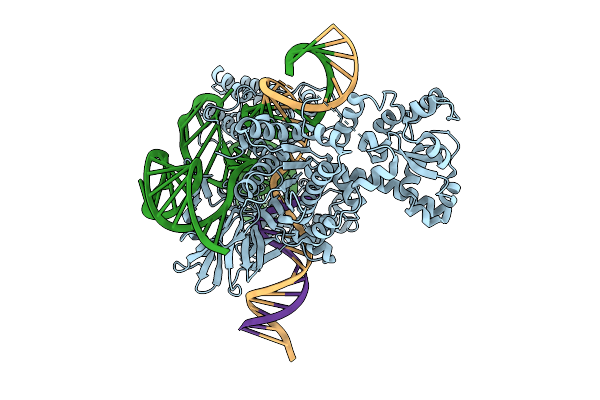
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In An Interrogation State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |
 |
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In An Interrogation State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |
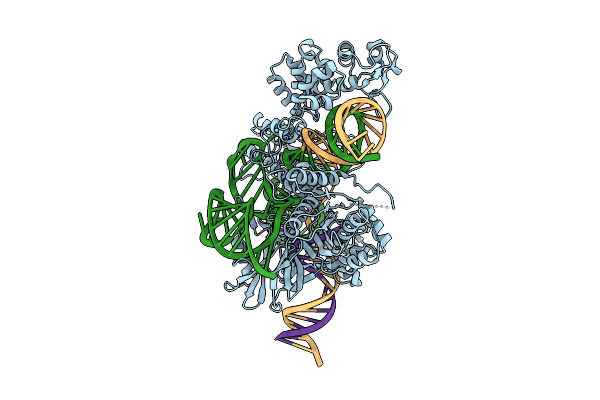 |
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In A Translocation State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: MG |
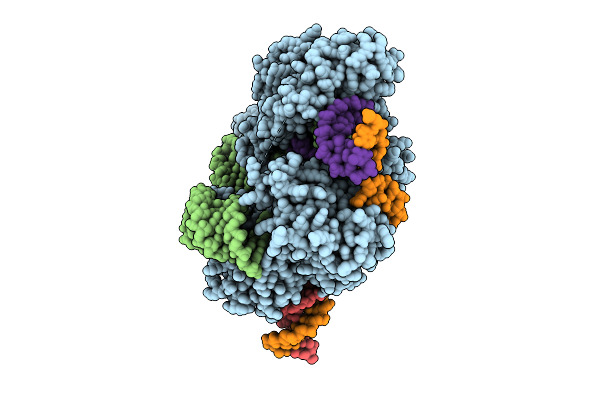 |
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In A Catalytically Active State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: K |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: DNA Ligands: HG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
Group Ii Intron Assembly Intermediate Domain 1, 2, 3 And 4 "Partly Open" State
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
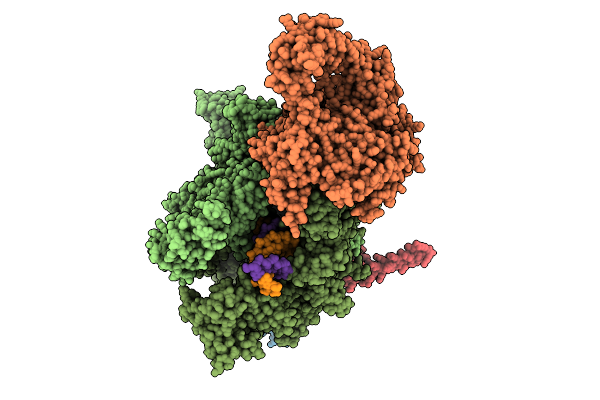 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
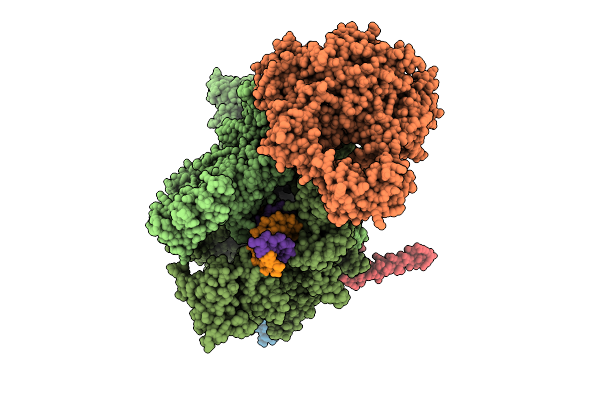 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
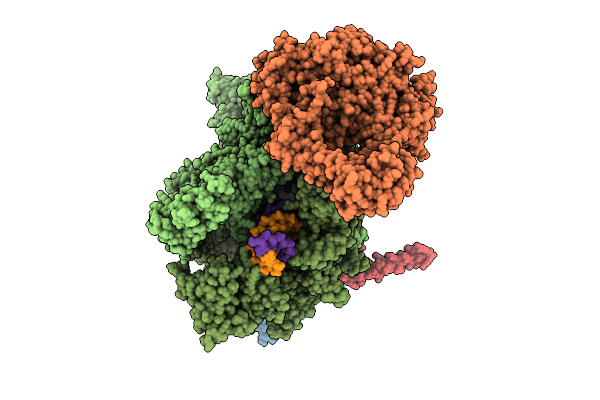 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
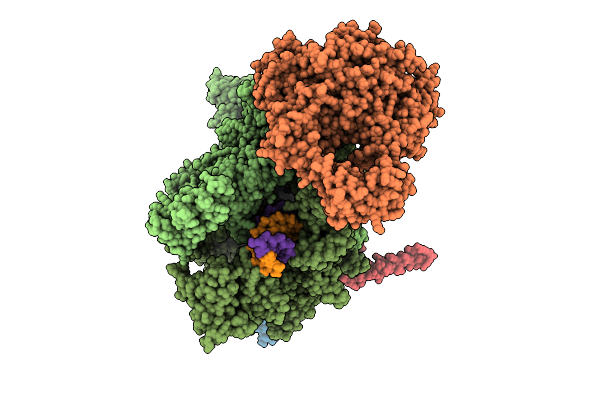 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: SO4, CL, TRS |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: ACY, PEG, NA |

