Search Count: 1,830
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HORMONE Ligands: CRS, SCN, ZN, GOL |
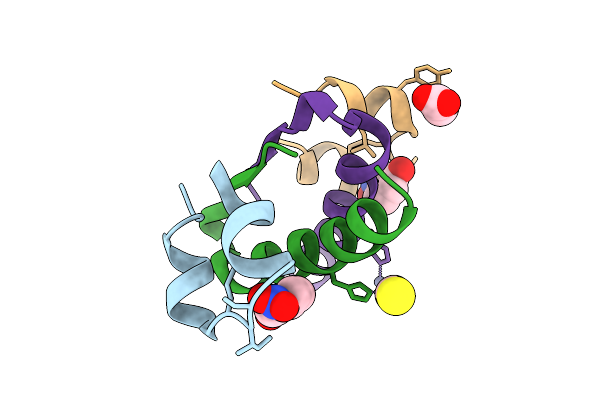 |
Rhombohedral Crystalline Form Of Human Insulin Complexed With M-Nitrophenol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HORMONE Ligands: ZCQ, SCN, ZN, ACT |
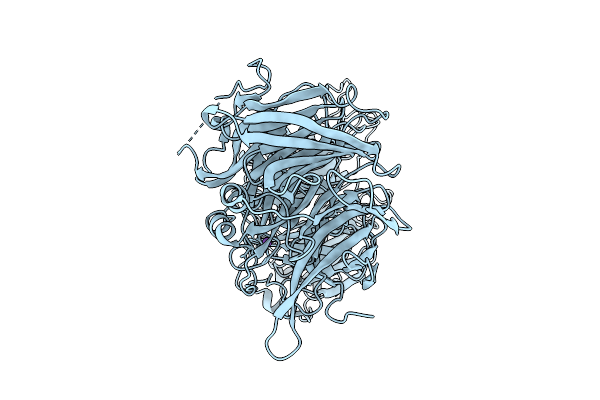 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: EXOCYTOSIS |
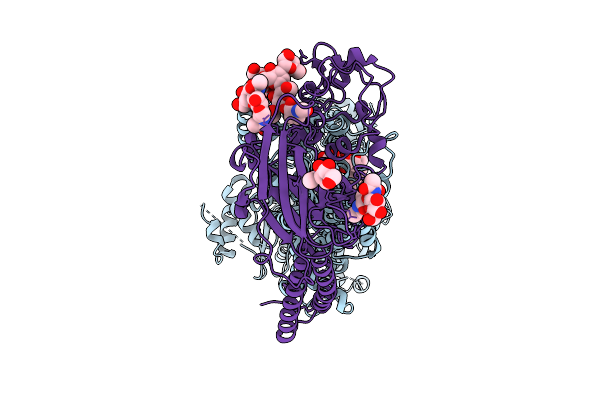 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
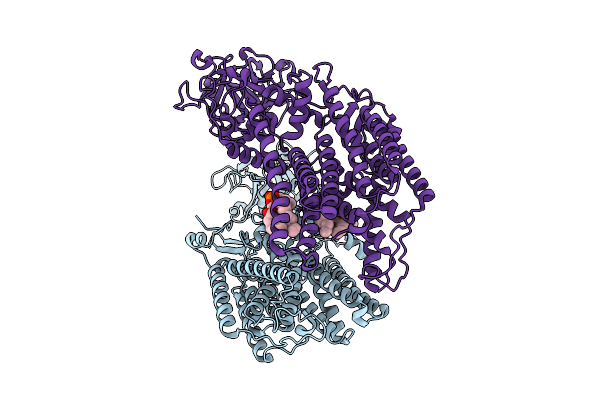 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: CPL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: NNM |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: CPL, MAN |
 |
Crystal Structure Of The Mir Domain Of The S. Cerevisiae Mannosyltransferase Pmt4 In Complex With Ccw5
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: PEG, GOL |
 |
Structure Of Human Adar2-R2D Complexed With Dsrna Containing 8-Azan And 2'-Deoxy-2'-Fluorouridine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: RNA BINDING PROTEIN/RNA Ligands: IHP, ZN, EDO, NA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MG, CL |
 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MG, CL |
 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MG, CL |
 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MG, CL, MES |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: SO4, MG, CL, MPD, MES |
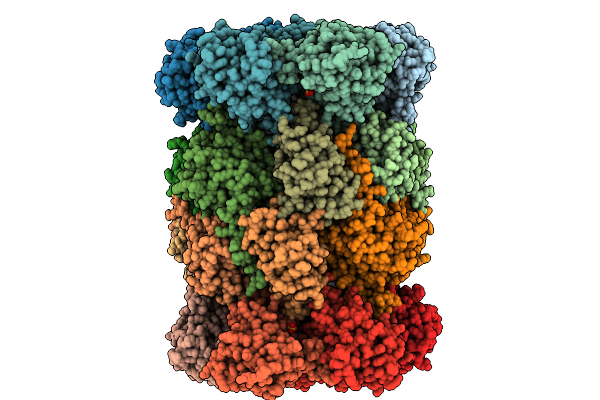 |
Yeast 20S Proteasome Mutant: Beta5_G128V (B5-Propeptide In Trans) In Complex With Mg132
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: SO4, MG, CL, MPD, MES |
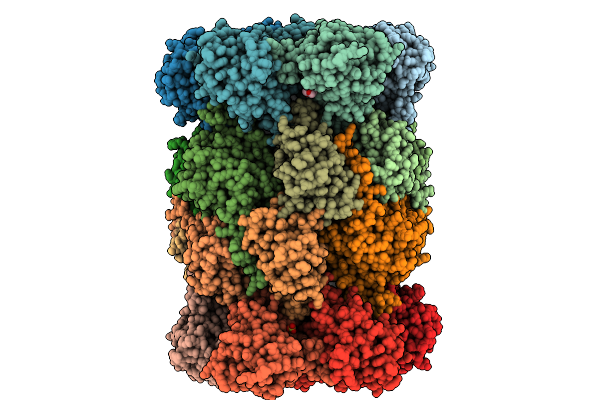 |
Yeast 20S Proteasome Mutant: Beta5_G128V (B5-Propeptide In Trans) In Complex With Carfilzomib
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MG, CL, MES, MPD, SO4 |
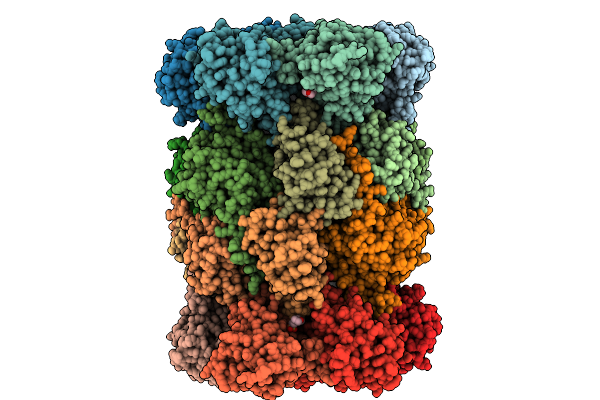 |
Yeast 20S Proteasome Mutant: Beta5_G128V (B5-Propeptide In Trans) In Complex With Onx0914
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MG, CL, MES, MPD |
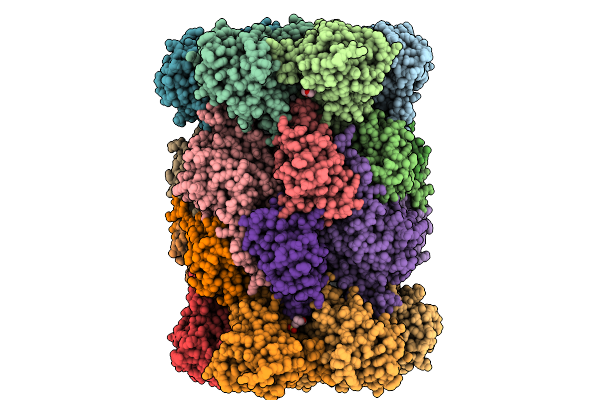 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MG, CL, MES, MPD |
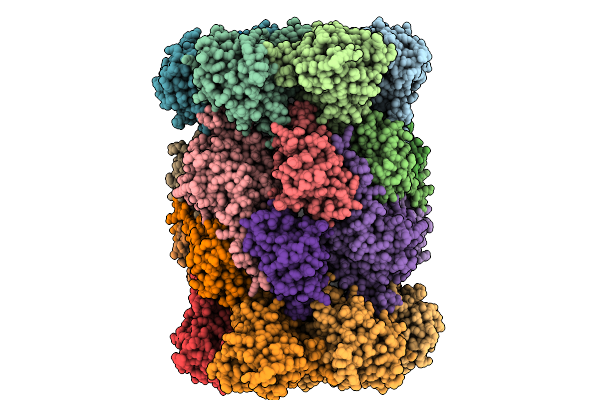 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MG, CL, MES |
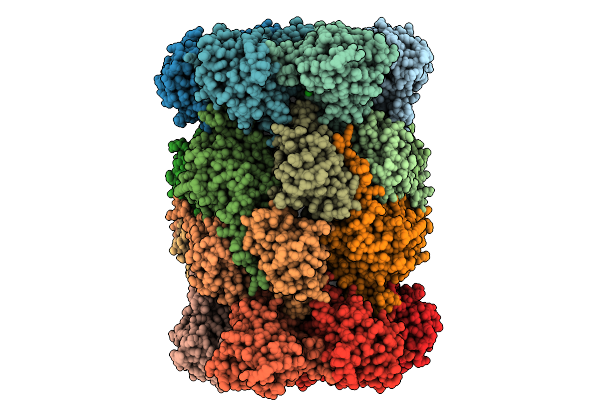 |
Yeast 20S Proteasome Mutant: Beta1_G128V (B1-Propeptide Deleted) In Complex With Mg132
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MG, CL, MES |

