Search Count: 503
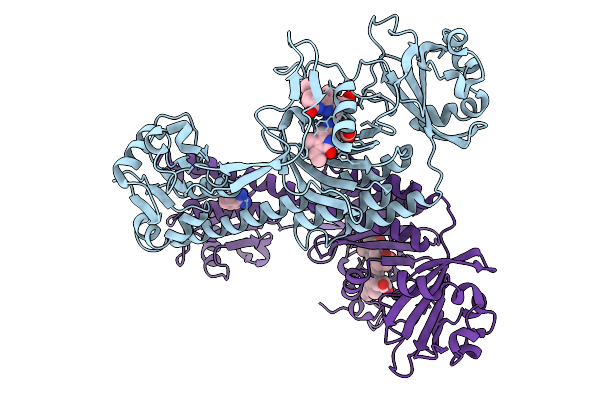 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 3 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: 3Q8, BEN |
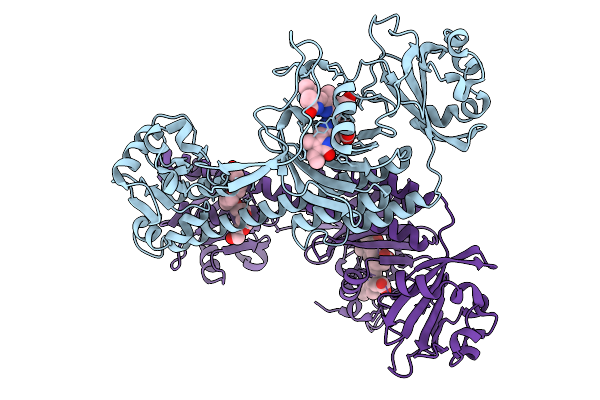 |
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
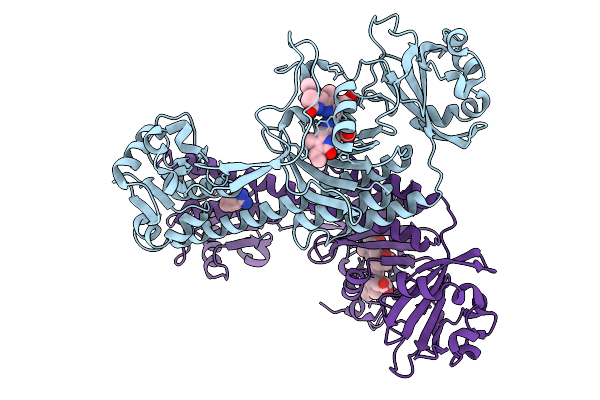 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 100 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
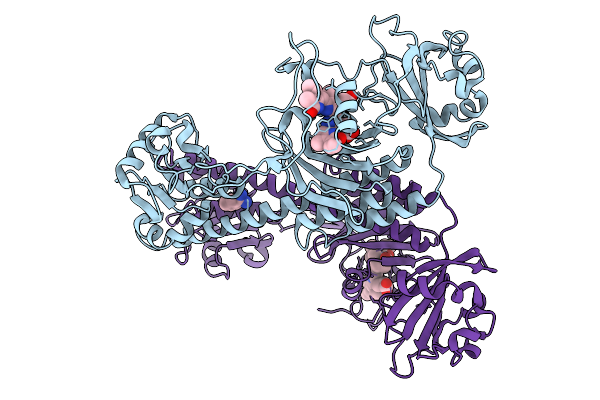 |
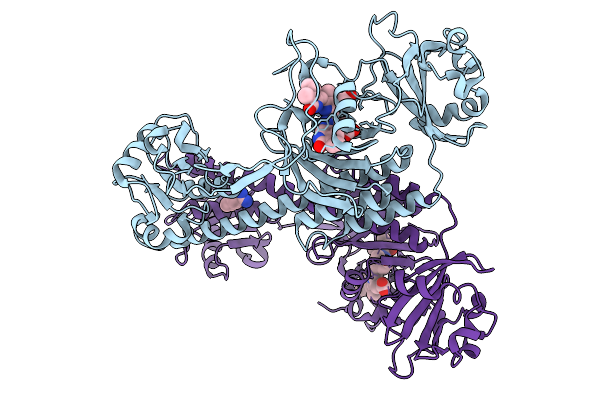
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
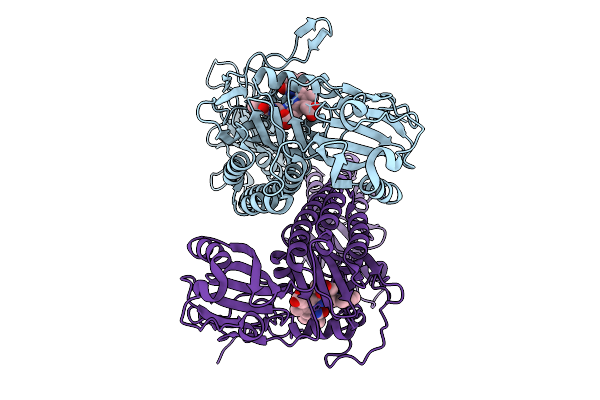
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
Prpfr Hybrid State Of The Pseudomonas Aeruginosa Bacteriophytochrome / Pabphp
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CYTOSOLIC PROTEIN Ligands: LBV |
 |
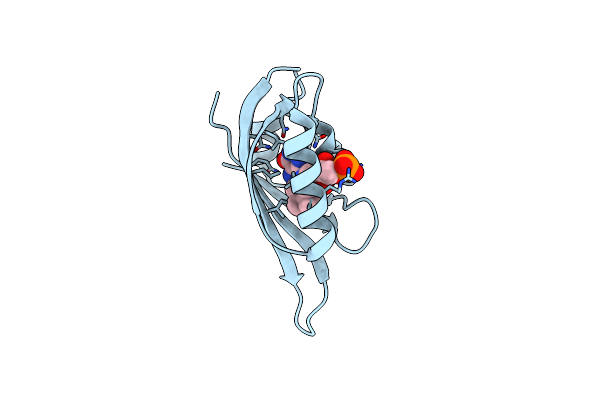
Structure Of Meiothermus Ruber Mrub_1259 Lov Domain With N- And C-Terminal Alpha Helices (Mrlove)
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: FLUORESCENT PROTEIN Ligands: FMN |
 |
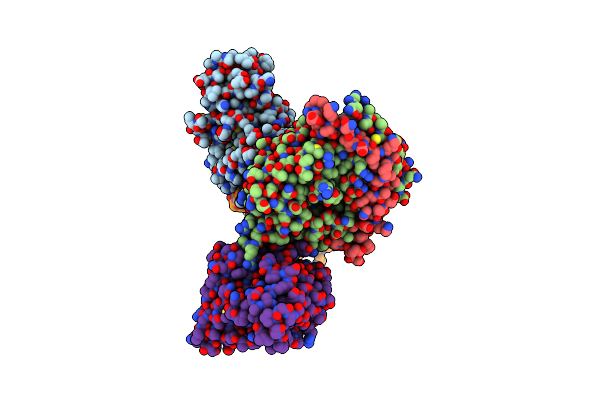
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: FLUORESCENT PROTEIN Ligands: FMN |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: STRUCTURAL PROTEIN Ligands: RET |
 |
Organism: Haloquadratum walsbyi dsm 16790
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: RET, MG |
 |
Organism: Chlamydomonas reinhardtii cc3269
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
 |
Sfx Structure Of Cracry 10 Ns After Photoexcitation Of The Oxidized Protein
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
 |
Sfx Structure Of Cracry 30 Ns After Photoexcitation Of The Oxidized Protein
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
 |
Sfx Structure Of Cracry 100 Ns After Photoexcitation Of The Oxidized Protein
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
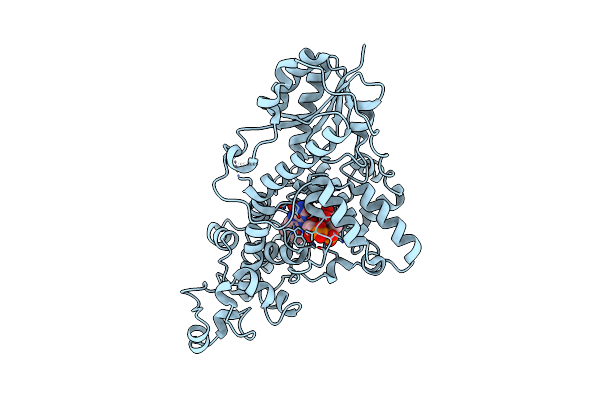 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
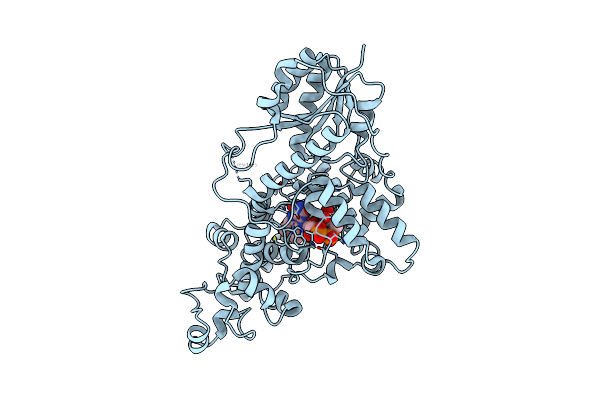 |
Sfx Structure Of Cracry 10 Us After Photoexcitation Of The Oxidized Protein
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
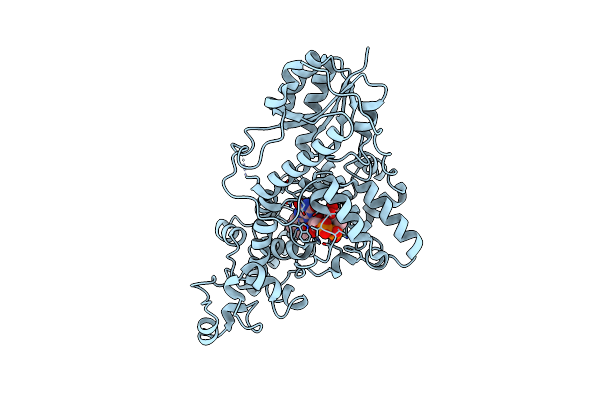 |
Sfx Structure Of Cracry 30 Us After Photoexcitation Of The Oxidized Protein
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
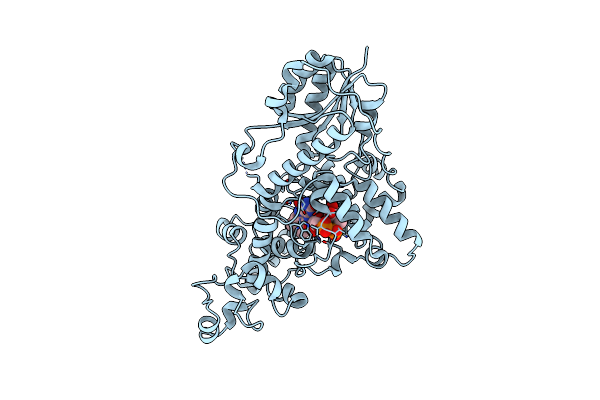 |
Sfx Structure Of Cracry 100 Us After Photoexcitation Of The Oxidized Protein
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
 |
Sfx Structure Of Cracry 300 Ns After Photoexcitation Of The Oxidized Protein
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |

